E2F5 Overexpression in Laryngeal Squamous Cell Carcinoma: Associations With Neutrophil Extracellular Traps in the Tumor Microenvironment
DOI:
https://doi.org/10.14740/wjon2610Keywords:
Laryngeal squamous cell carcinoma, E2F5, Neutrophil extracellular traps, Tumor microenvironment, Single-cell RNA sequencingAbstract
Background: Laryngeal squamous cell carcinoma (LSCC) is a common malignant tumor of the head and neck, associated with smoking and excessive alcohol consumption. The objective was to investigate the expression pattern of E2F transcription factor 5 (E2F5) in LSCC and its association with neutrophil extracellular traps (NETs), elucidating its role in the tumor microenvironment.
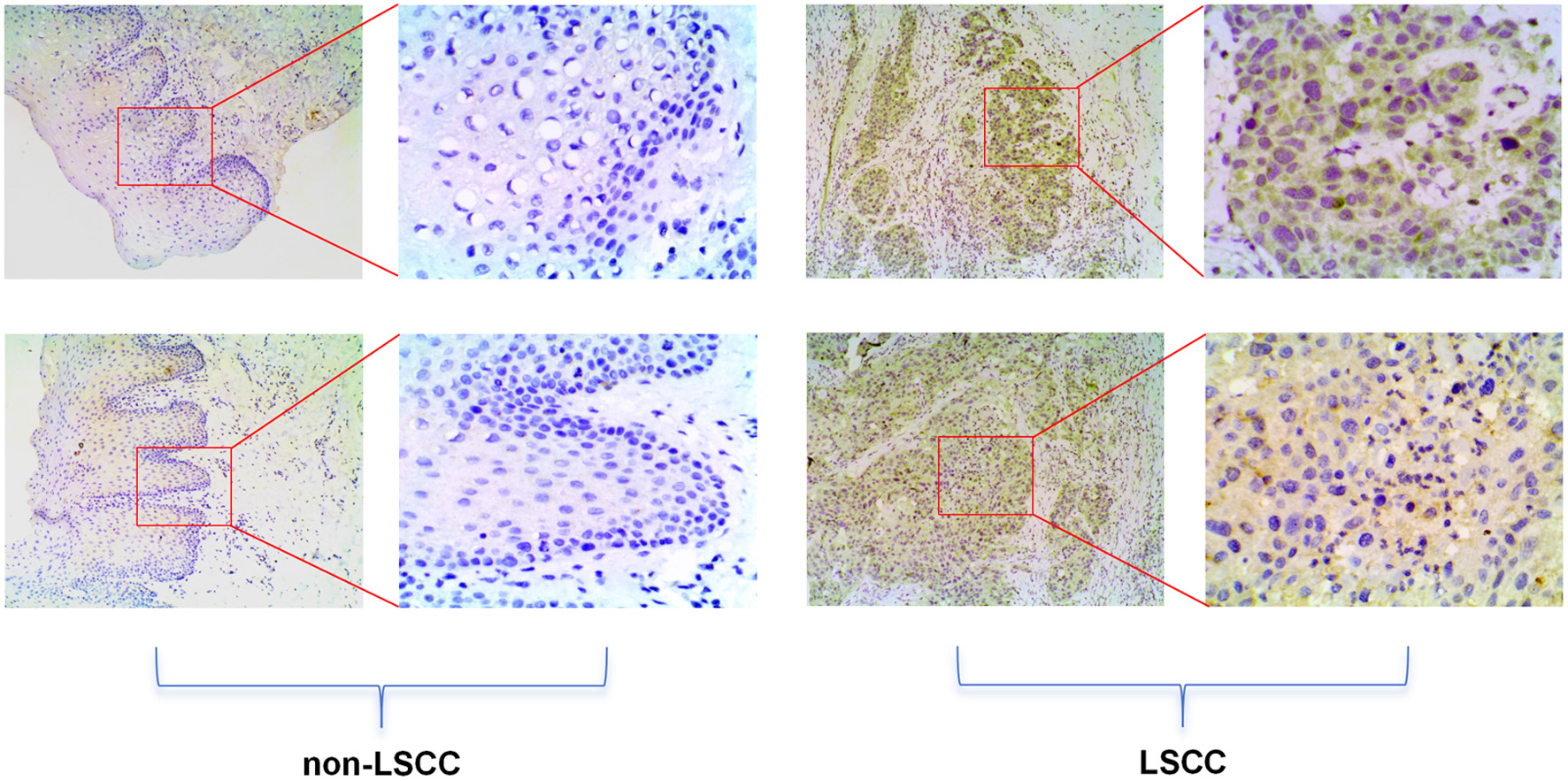
Methods: At the cellular level, single-cell RNA sequencing (scRNA-seq) was employed to analyze the expression of E2F5 and NETs-related genes (S100A8, S100A9, LCN2, etc.). At the tissue level, spatial transcriptomics (ST) was used to examine the E2F5 expression pattern. At the mRNA level, E2F5 expression was assessed through mRNA expression profiling, and at the protein level, expression was validated using immunohistochemistry (IHC) on tissue specimens, including 10 LSCC cases (laryngeal, hypopharyngeal, and oropharyngeal squamous cell carcinomas) and 10 non-LSCC controls (benign lesions such as mucoceles, hemangiomas, and polyps). Clustered regularly interspaced short palindromic repeats (CRISPR) knockout screening combined with the CERES algorithm was utilized to evaluate the impact of E2F5 on LSCC cell line proliferation, with negative/positive dependency scores indicating suppression/promotion of growth, respectively. Single-sample Gene Set Enrichment Analysis (ssGSEA) was used to analyze the correlation between E2F5 and immune cells, and chromatin immunoprecipitation sequencing (ChIP-seq) was performed to validate the transcriptional regulation of NETs-related genes by E2F5. Statistical analyses included Wilcoxon, standardized mean difference (SMD), receiver operating characteristic (ROC), and summary receiver operating characteristic (sROC).
Results: E2F5 exhibited high expression in LSCC epithelial cells and tissues, with elevated expression at both mRNA and protein levels (SMD = 0.24, 95% confidence interval (CI) = 0.0309 - 0.448, sROC area under the curve (AUC) = 0.71, IHC P = 7.2 × 10-6, ROC AUC = 1). Knockdown of E2F5 significantly inhibited proliferation in LSCC cell lines (e.g., BICR31, BICR16) (inhibition score < 0). High E2F5 expression was positively correlated with T-helper cells and natural killer (NK) CD56bright cells (R = 0.251, 0.175, P < 0.05) and negatively correlated with neutrophils and Th17 cells (R = -0.293, -0.260, P < 0.05). Cellular and tissue-level analyses revealed high NETs expression in LSCC, with E2F5 also highly expressed in NETs-related cells and regions. ChIP-seq analysis confirmed that E2F5 regulates NETs-related genes. Functional enrichment analysis indicated that E2F5-related genes are involved in transcriptional regulation, chromatin organization, and immune regulation.
Conclusion: E2F5 is highly expressed in LSCC and is associated with the regulation of NETs-related genes. It may contribute to tumor proliferation and immune evasion by reshaping the tumor microenvironment, highlighting E2F5 as a potential therapeutic target that warrants further functional validation.
Published
Issue
Section
License
Copyright (c) 2025 The authors

This work is licensed under a Creative Commons Attribution 4.0 International License.









