Metabolic Reprogramming-Related Genes in Lung Adenocarcinoma: Identification and Prognostic Model Construction
DOI:
https://doi.org/10.14740/wjon2604Keywords:
Lung adenocarcinoma, Metabolic reprogramming genes, Weighted gene co-expression network analysis, Consensus clustering, Prognostic modelAbstract
Background: Lung adenocarcinoma (LUAD), the predominant histological subtype of lung cancer, persists in presenting a dismally low 5-year overall survival (OS) rate, notwithstanding advancements in treatment modalities. There exists a pressing necessity for the identification of innovative biomarkers that can enhance prognostic assessments and facilitate individualized therapeutic strategies. The objective of this investigation was to clarify the involvement of genes associated with metabolic reprogramming in the progression of LUAD and to evaluate their viability as prognostic indicators.
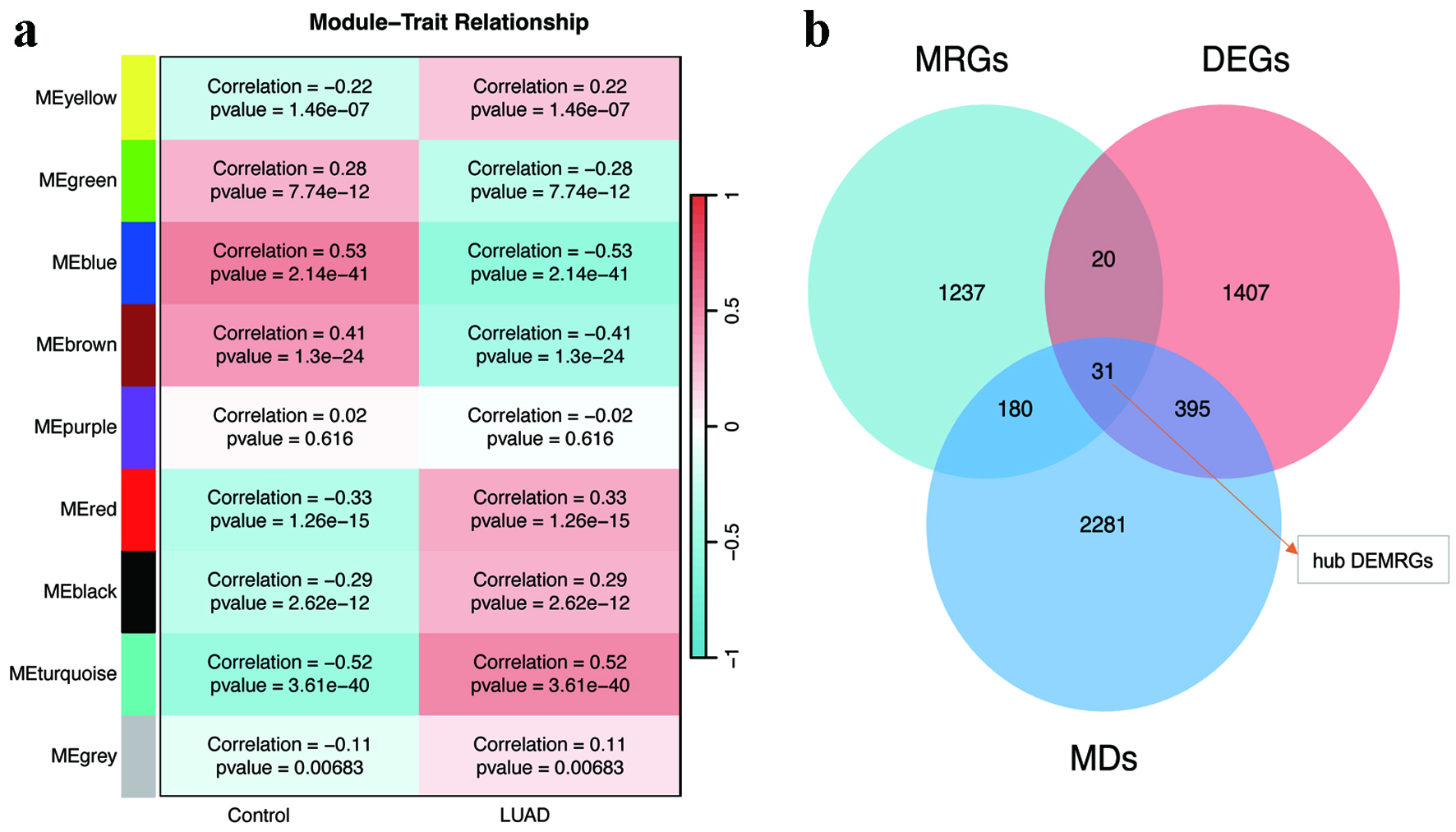
Methods: An analysis of differential gene expression was performed utilizing The Cancer Genome Atlas (TCGA)-LUAD dataset, supplemented by a weighted gene co-expression network analysis (WGCNA). Through intersection analysis focusing on metabolic reprogramming genes (MRGs), pivotal differentially expressed metabolic reprogramming genes (hub DEMRGs) were identified. Consensus clustering categorized patients into subtypes based on these genes. Functional enrichment analysis and immune microenvironment characterization were conducted, followed by Cox and least absolute shrinkage and selection operator (LASSO) regression analyses to construct a prognostic risk model.
Results: A total of 31 hub DEMRGs were identified. Patients were classified into two distinct subtypes (C1 and C2), with the C2 subtype exhibiting a markedly reduced OS rate. Functional enrichment revealed significant activation of nuclear division and cell cycle pathways in C2. Immune profiling demonstrated an immunosuppressive phenotype in C2, characterized by elevated M2 macrophage infiltration and reduced CD8+ T cells. The risk model based on five critical hub DEMRGs showed robust predictive performance (area under the curve (AUC): 0.68 - 0.71), and high-risk patients displayed unique immune cell infiltration patterns.
Conclusions: This research highlights the critical role of MRGs in LUAD prognosis and their potential for clinical application. The identified subtypes and risk model provide insights into tumor heterogeneity and immunosuppressive mechanisms, offering potential targets for individualized therapy.
Published
Issue
Section
License
Copyright (c) 2025 The authors

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.









