| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 16, Number 5, October 2025, pages 487-501
Targeting the Phosphoinositide 3-Kinase/Protein Kinase B Pathway Suppresses Y-Box Binding Protein 1 Expression and Inhibits Colorectal Cancer Progression
Hui Shana, c, Yan Wanga, c, Siyu Hua, Yuting Wanga, Rong Qina, Niu Zhanga, Guangyu Tianb, d, Zhiyuan Qiua, d
aDepartment of Oncology, Zhenjiang First People’s Hospital, Zhenjiang, Jiangsu, China
bDepartment of Oncology, Jiangdu People’s Hospital Affiliated to Yangzhou University, Yangzhou, Jiangsu, China
cThese authors contributed equally to this work.
dCorresponding Authors: Guangyu Tian, Department of Oncology, Jiangdu People’s Hospital Affiliated to Yangzhou University, Yangzhou, Jiangsu, China. Zhiyuan Qiu, Department of Oncology, Zhenjiang First People’s Hospital, Zhenjiang, Jiangsu, China
Manuscript submitted July 9, 2025, accepted August 22, 2025, published online September 17, 2025
Short title: Targeting PI3K/AKT Lowers YBX1 in CRC
doi: https://doi.org/10.14740/wjon2640
| Abstract | ▴Top |
Background: Colorectal cancer (CRC) is one of the most prevalent and lethal malignancies worldwide, often characterized by the aberrant activation of multiple signaling pathways. Y-box binding protein 1 (YBX1), a multifunctional regulator of transcription and translation, has been identified as an oncogenic factor in various solid tumors. However, its expression profile and mechanistic role in CRC remain largely unclear.
Methods: In this study, integrative bioinformatic analyses were conducted on The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets to assess YBX1 expression and its correlation with CRC progression. Functional assays, including cell proliferation and migration assays, were performed to investigate the role of YBX1 in CRC cells. The impact of YBX1 on the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) signaling pathway was evaluated, and the effects of the PI3K inhibitor buparlisib (BKM120) on YBX1-driven cellular phenotypes were also tested.
Results: YBX1 was found to be significantly upregulated in CRC tissues and was closely associated with the activation of the PI3K/AKT signaling pathway. YBX1 overexpression promoted CRC cell proliferation and migration, whereas knockdown of YBX1 inhibited these processes. Mechanistically, YBX1 was shown to enhance PI3K/AKT signaling activity, promoting malignant phenotypes in CRC. Treatment with BKM120 partially reversed these effects. Additionally, Gene Set Enrichment Analysis (GSEA) identified enrichment of reactive oxygen species (ROS)-related pathways in YBX1-high CRC samples.
Conclusions: This study highlights the oncogenic role of YBX1 in CRC and reveals a potential YBX1-PI3K/AKT regulatory axis that may serve as a promising therapeutic target. The findings suggest that targeting this axis could provide a novel strategy for CRC treatment, especially under hypoxic or microenvironmental stress conditions.
Keywords: Colorectal cancer; YBX1; PI3K/AKT signaling pathway; BKM120; Hypoxia
| Introduction | ▴Top |
Colorectal cancer (CRC) is the third most common malignancy worldwide and one of the leading causes of cancer-related mortality [1]. According to the latest data from the International Agency for Research on Cancer (IARC), the incidence and mortality rates of CRC continue to rise globally, particularly in developing countries, where early detection and effective treatment remain major challenges [2]. Although current therapeutic strategies, including surgery, radiotherapy, and chemotherapy, have improved patient survival to some extent, their efficacy in advanced or metastatic CRC is still limited, and long-term prognosis remains poor [3]. Therefore, elucidating the molecular mechanisms underlying CRC progression and identifying novel therapeutic targets is of great clinical and scientific importance.
Y-box binding protein 1 (YBX1) is a highly conserved nucleic acid-binding protein that plays dual roles in the regulation of transcription and translation. It plays a role in various essential biological processes, including DNA repair, mRNA splicing, and protein synthesis [4, 5]. Previous studies have demonstrated that YBX1 is overexpressed in a variety of solid tumors and plays a crucial role in promoting tumor cell proliferation, migration, invasion, and drug resistance. Its oncogenic role has been well documented in breast cancer [6, 7], gastric cancer [8], and non-small cell lung cancer (NSCLC) [9]. However, the expression characteristics, functional roles, and upstream regulatory mechanisms of YBX1 in CRC remain largely unexplored.
In this study, we analyzed The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets and found that YBX1 is significantly upregulated in CRC tissues and is closely associated with tumor differentiation grade [10]. Using a combination of in vitro functional assays, RNA sequencing, and pharmacological inhibition with the phosphoinositide 3-kinase (PI3K) pathway inhibitor buparlisib (BKM120), we systematically investigated the oncogenic role of YBX1 and its underlying regulatory mechanisms in CRC. Additionally, we explored the regulatory characteristics of YBX1 under conditions of reactive oxygen species (ROS) enrichment and hypoxic conditions, and preliminarily assessed its potential impact on the tumor immune microenvironment [11]. This study aims to elucidate the role of the PI3K/protein kinase B (AKT)-YBX1 regulatory axis in CRC and to provide a theoretical basis and potential strategy for targeted therapy in CRC.
| Materials and Methods | ▴Top |
Data and resources
The CRC-related transcriptomic data used in this study were obtained primarily from the GEO and TCGA database. Five public datasets, including GSE21815, GSE31905, GSE35279, GSE41657, and GSE81558, were selected from the GEO database to analyze the expression characteristics of YBX1 in CRC and its correlation with clinical indicators. These datasets were obtained through the official GEO platform and cover expression data of both tumor tissues and normal tissues from CRC patients. Additionally, the study accessed and analyzed the expression data of CRC (COAD and READ) from the TCGA database using the Gene Expression Profiling Interactive Analysis (GEPIA) online tool, in order to further validate the expression trends of YBX1 in different samples and its correlation with survival prognosis.
For functional enrichment analysis, Gene Set Enrichment Analysis (GSEA) software was used to perform pathway enrichment analysis on differentially expressed genes in the TCGA samples of YBX1 high and low expression groups. Pathways potentially related to the function of YBX1 were identified, providing a reference for subsequent mechanistic studies. All the data and analysis tools used were publicly available and comply with the relevant usage guidelines.
All experiments were conducted using established cell lines under standard laboratory conditions. The study did not involve human participants, identifiable personal data, or animal experimentation. Hence, ethical approval was not required, in accordance with institutional and journal policies.
Cell culture and treatments
This study used six cell lines, including five human CRC cell lines (Caco-2, HCT 116, HT-29, SW480, and SW620) and one mouse fibroblast cell line (L929). The Caco-2 cells were cultured in high-glucose Dulbecco’s Modified Eagle Medium (DMEM, Gibco) containing 20% fetal bovine serum (FBS, Gibco); HCT 116 and HT-29 cells were cultured in McCoy’s 5A medium (Gibco) with 10% FBS; SW480 and SW620 cells were cultured in high-glucose DMEM containing 10% FBS. L929 cells were cultured in high-glucose DMEM with 10% FBS. All cells were maintained in a 37 °C, 5% CO2 incubator, and the culture medium was changed every 1 - 2 days.
For the drug treatment experiments, HCT 116, HT-29, SW480, and SW620 cells, as well as stable cell lines with interference or overexpression of YBX1, were treated with 300 µM cobalt chloride (CoCl2, Sigma) for 24 h to simulate hypoxic conditions. These four CRC cell lines were also treated with BKM120 (Selleck) for 24 h at their respective half-maximal inhibitory concentration (IC50) concentrations. As a normal control, L929 cells were not treated with CoCl2 but were treated with 2, 5, and 8 µM BKM120 to evaluate the potential effects of the drug on normal cell viability.
RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction (qRT-PCR)
The CRC cell lines used in this study include Caco-2, HCT 116, HT-29, SW480, and SW620. Once the cells reached an appropriate density, total RNA was extracted using Trizol reagent (Takara Bio, Japan) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA), and equal amounts of RNA were used to synthesize cDNA using the PrimeScript RT reagent kit (Takara, Japan).
For qRT-PCR, 2× SYBR Green premix (Takara, Japan) was used, and reactions were performed on a 7500 Real-Time PCR system (Applied Biosystems, USA). The primer sequences for target genes are as follows: 18sRNA (forward: 5'-TGCGAGTACTCAACACCAACA-3', reverse: 5'-GCATATCTTCGGCCCACA-3'), YBX1 (forward: 5'-TAGACGCTATCCACGTCGTAG-3', reverse: 5'-ATCCCTCGTTCTTTTCCCCAC-3'), hypoxia-inducible factor 1-alpha (HIF-1α) (forward: 5'-GAACGTCGAAAAGAAAAGTCTCG-3', reverse: 5'-CCTTATCAAGATGCGAACTCACA-3'). In the experiment, 18S rRNA was used as an internal reference gene, and the relative gene expression levels were calculated using the 2-ΔΔCt method.
Western blot assay
Total protein was extracted from cells using RIPA lysis buffer (P0013B, Beyotime, Shanghai, China) containing protease and phosphatase inhibitors (ab201119, Abcam, Shanghai, China). The protein concentration was determined using a BCA protein assay kit (Pierce Biotechnology, USA) according to the manufacturer’s instructions. Equal amounts of protein were separated by SDS-PAGE and transferred to a nitrocellulose (NC) membrane (BioTrace™ NT, PALL). The membrane was blocked with 5% non-fat dry milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4 °C. The primary antibodies used in this study include: YBX1 (1:10,000, Proteintech, 20339-1-AP), HIF-1α (1:1,000, Cell Signaling Technology, #36169), AKT (1:2,000, Proteintech, 10176-2-AP), p-AKT (Ser473) (1:2,000, Proteintech, 66444-1-Ig), and GAPDH (1:10,000, Proteintech, 10494-1-AP). The next day, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature, and protein bands were detected using an ECL chemiluminescent reagent kit.
Western blot densitometric analysis
Western blot band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA). For each blot, the intensity of the target protein bands (e.g., HIF-1α) was normalized to the corresponding GAPDH band to correct for loading variations. Relative expression levels were calculated by setting the control group to 1.0, and values from at least three independent experiments were averaged. The results are presented as mean ± standard deviation (SD). Statistical significance was determined using Student’s t-test (for two groups) or one-way analysis of variance (ANOVA) (for multiple groups), with P < 0.05 considered statistically significant.
Lentiviral transduction
To overexpress YBX1, lentiviral vectors carrying the YBX1 gene were purchased from Heyuan Bio (Shanghai, China) HT-29 and SW480 cells were then infected with these vectors according to the manufacturer’s instructions. After infection, cells were selected with 10 µg/mL blasticidin for 2 weeks to establish stable YBX1 overexpressing cell lines. To knock down YBX1, lentiviral vectors carrying shRNA sequences targeting YBX1 were used to infect HCT 116 and SW620 cells. The specific shRNA sequences are as follows: shYBX1-1: 5'-CCAGTTCAAGGCAGTAAATAT-3'; shYBX1-2: 5'-AGCAGACCGTAACCATTATAG-3'; shYBX1-3: 5'-GACGGCAATGAAGAAGATAAA-3'. After infection, cells were selected with 5 µg/mL puromycin for 2 weeks to establish stable YBX1 knockdown cell lines. The efficiency of YBX1 overexpression or knockdown in each cell line was verified by qRT-PCR and Western blot.
siRNA transfection
YBX1 targeting siRNA and negative control siRNA were purchased from GenePharm (Shanghai, China) for transfection into HCT 116 and SW620 CRC cells. The transfection was performed according to the manufacturer’s instructions. Cells were collected 48 h after transfection for siRNA efficiency validation and subsequent experiments. The antisense sequences for the siRNAs used are as follows: siYBX1-1 (YBX1-Homo-524): 5'-UAUCCGUUCCUUACAUUGAAC-3'; siYBX1-2 (YBX1-Homo-620): 5'-CCUACACUGCGAAGGUACUUC-3'.
Cell proliferation and colony formation assay
Cell proliferation was assessed using the cell counting kit-8 (CCK-8) method. Cells (1 - 2 × 103) were seeded in a 96-well plate, with three replicates per group, and cultured for 5 days. At the set time points, the culture medium was removed, and 100 µL of CCK-8 reagent (diluted at 1:10 with culture medium) was added to each well, incubated for 60 min at 37 °C, and the absorbance (optical density (OD) value) was measured at 450 nm using a multifunctional microplate reader (Bio-Rad, USA) to reflect cell activity.
For colony formation assays, 1 × 103 cells were seeded in a 6-cm dish and cultured for 10 - 14 days until visible colonies formed. The colonies were washed twice with 1× phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min, and stained with 0.2% crystal violet for 1 h. After washing off excess dye and air-drying, the number of colonies with a diameter greater than 100 µm was counted to evaluate the cell colony-forming ability.
Transwell migration assay
The cell migration assay was performed using Transwell chambers with an 8-µm pore size (JET BOIFIL, Guangzhou, China). Cells (5 × 104) were suspended in 200 µL serum-free medium and seeded into the upper chamber of the Transwell. The lower chamber was filled with 800 µL medium containing 10% FBS as a chemotactic agent. After incubation for 24 h at 37 °C, 5% CO2, the non-migrated cells in the upper chamber were gently removed with a cotton swab. The migrated cells were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet for 30 min, washed with PBS, and randomly counted in five fields of view under an inverted microscope to assess cell migration ability.
Immunofluorescence (IF) staining
Bone marrow-derived macrophages (BMDMs) were isolated from C57BL/6 mice and differentiated. The cells were then seeded onto sterile coverslips and incubated in a 37 °C, 5% CO2 incubator for 24 h. Cells were treated with different concentrations of BKM120 (2, 5, and 8 µM) for 24 h, and the control group was treated with lipopolysaccharide (LPS, 1 µg/mL).
After treatment, cells were washed twice with 1× PBS, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 at room temperature for 10 min. Non-specific binding sites were blocked with 5% bovine serum albumin (BSA) for 1 h, followed by overnight incubation with primary antibodies at 4 °C. Primary antibodies included anti-iNOS and anti-Arg-1 antibodies (both purchased from Abclonal), used to detect M1 and M2 macrophage polarization. On the following day, the cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG (green) secondary antibody for 1 h at room temperature and stained with DAPI (blue) to label cell nuclei. Cytoskeletal structure was stained with fluorescein isothiocyanate (FITC)-labeled phalloidin (Biosharp, green). All images were captured using an inverted fluorescence microscope, and fluorescence signals were observed and quantified using ImageJ software.
Quantification was performed on ≥ 5 random fields per condition from ≥ 3 independent biological replicates. For each field, mean fluorescence intensity (MFI) of induced nitric oxide synthase (iNOS) and Arg-1 was measured with identical regions of interest (ROIs) and local background subtraction in ImageJ; values were normalized within each experiment to the LPS + vehicle group (set to 1.0). Data are presented as mean ± SD and were analyzed by two-tailed Student’s t-test (two groups) or one-way ANOVA with Tukey’s post hoc test (multiple doses); P < 0.05 was considered statistically significant.
Live/dead cell staining assay
To evaluate the toxicity of the PI3K/AKT pathway inhibitor BKM120 on normal cells, L929 mouse fibroblast cells underwent live/dead cell staining using Calcein-AM/PI dual staining. Cells were seeded in six-well plates at a density of 2 × 104 cells per well and incubated for 24 h at 37 °C and 5% CO2. After attachment, cells were treated with different concentrations of BKM120 (0, 2, 5, and 8 µM) for 24 h.
After treatment, the culture medium was discarded, and cells were washed twice with PBS. Aworking solution of Calcein-AM and propidium iodide (PI) working solution (Biosharp, China) was added according to the manufacturer’s instructions, after which the cells were incubated in the dark at room temperature for 15 min. After staining, the cells were observed under an inverted fluorescence microscope, and images were collected. Green fluorescence (Calcein-AM) represents live cells, and red fluorescence (PI) represents dead cells. Image analysis was performed using ImageJ software to quantify the proportion of dead cells in different treatment groups and evaluate the toxicity of BKM120 on L929 normal cells.
| Results | ▴Top |
YBX1 high expression in CRC and its association with the PI3K/AKT pathway
To investigate the expression characteristics of YBX1 in CRC, we first conducted bioinformatics analyses using TCGA and GEO datasets, including GSE21815, GSE31905, GSE35279, and GSE44076. The results showed that YBX1 was significantly upregulated in both colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) tissues compared with normal colorectal tissues (Fig. 1c, e).
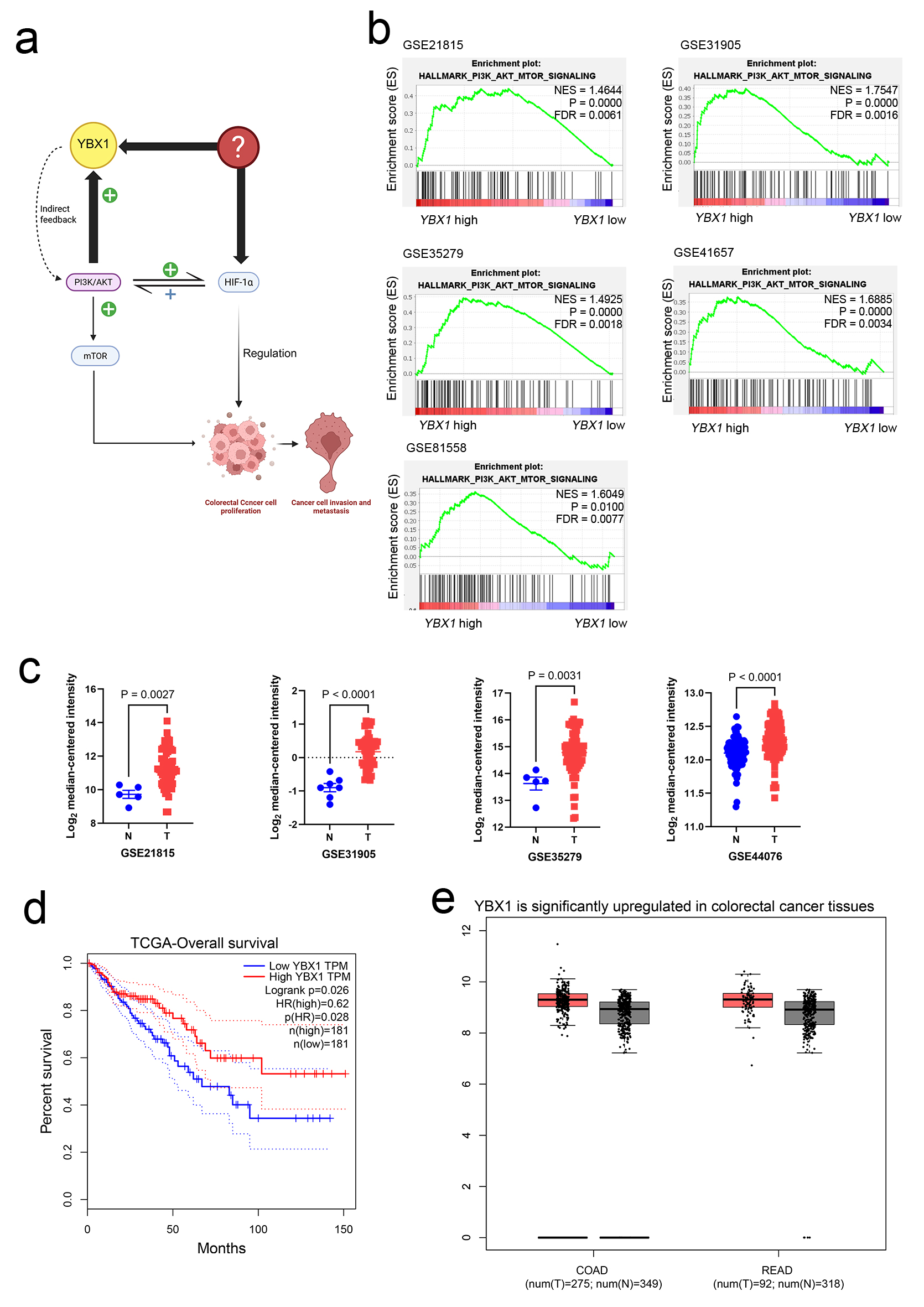 Click for large image | Figure 1. YBX1 high expression in CRC and its association with the PI3K/AKT pathway. (a) Schematic diagram showing the potential mechanism of YBX1 in CRC. (b) GSEA showing significant enrichment of the PI3K/AKT/mTOR signaling pathway in YBX1 high-expression samples. (c) GEO database analysis (GSE21815, GSE31905, GSE35279, and GSE44076) showing significant upregulation of YBX1 expression in COAD and READ tissues. (d) TCGA database analysis showing that high YBX1 expression is associated with poor survival prognosis in CRC patients. (e) TCGA data further validating the high expression of YBX1 in COAD and READ tumor tissues. AKT: protein kinase B; COAD: colon adenocarcinoma; CRC: colorectal cancer; GEO: Gene Expression Omnibus; GSEA: Gene Set Enrichment Analysis; mTOR: mammalian target of rapamycin; PI3K: phosphoinositide 3-kinase; READ: rectum adenocarcinoma; TCGA: The Cancer Genome Atlas; YBX1: Y-box binding protein 1. |
Survival analysis indicated that high YBX1 expression was closely associated with lower tumor differentiation grade and poorer overall prognosis (Fig. 1d). Furthermore, GSEA revealed that the PI3K/AKT signaling pathway was significantly enriched in samples with high YBX1 expression (Fig. 1b), suggesting a potential mechanistic link between YBX1 and PI3K/AKT pathway activation in CRC.
Taken together, these findings collectively suggest that YBX1 is upregulated in CRC and may contribute to tumor progression via activation of the PI3K/AKT signaling pathway, thereby providing a mechanistic basis for further functional validation and therapeutic exploration.
YBX1 regulates proliferation and colony formation of CRC cells
To assess the functional role of YBX1, we initially examined its expression across five CRC cell lines (Fig. 2a, Supplementary Material 1A, wjon.elmerpub.com). YBX1 expression was found to be relatively high in HCT 116 and SW620 cells, while lower levels were observed in HT-29 and SW480 cells. Based on these results, we constructed YBX1 knockdown and overexpression models (Fig. 2b, Supplementary Material 1B, C, wjon.elmerpub.com). CCK-8 assays showed that YBX1 knockdown significantly inhibited cell proliferation, while YBX1 overexpression promoted cell proliferation (Fig. 2c, d). Colony formation assays further confirmed that YBX1 expression levels were closely related to colony formation capacity, with knockdown significantly reducing colony number and overexpression significantly enhancing it (Fig. 2c, d).
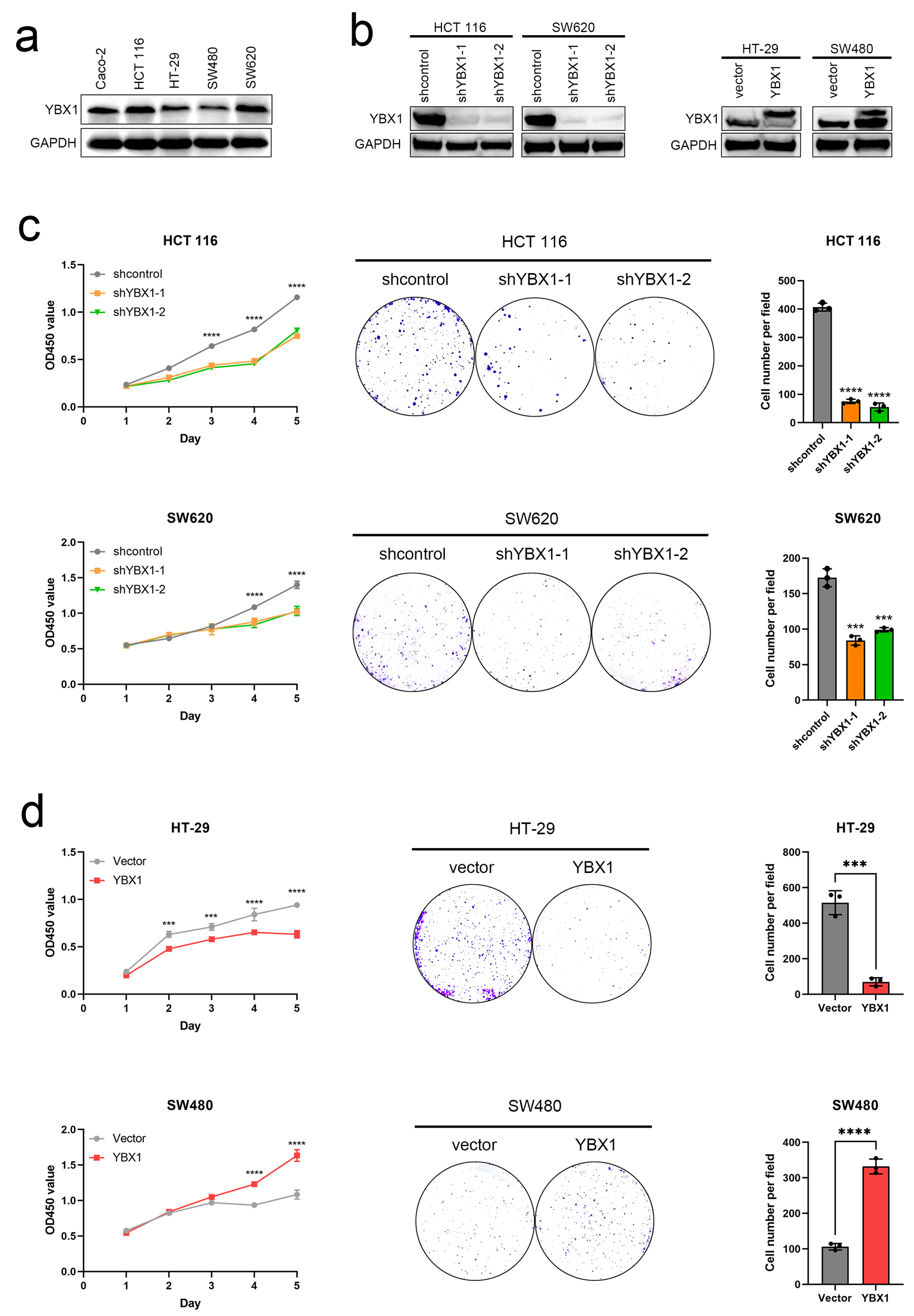 Click for large image | Figure 2. YBX1 regulates CRC cell proliferation and colony formation. (a) Expression levels of YBX1 in five CRC cell lines. (b) Western blot verification of YBX1 knockdown in HCT 116 and SW620 cells and YBX1 overexpression in HT-29 and SW480 cells. (c) CCK-8 assays showing that YBX1 knockdown inhibits cell proliferation, while overexpression promotes cell proliferation. (d) Colony formation assays showing that YBX1 knockdown decreases colony formation ability, while overexpression enhances colony formation ability. CCK-8: cell counting kit-8; CRC: colorectal cancer; YBX1: Y-box binding protein 1. |
YBX1 promotes CRC cell migration
Transwell migration assays further confirmed that YBX1 significantly enhanced CRC cell migration ability, suggesting that it plays a key role in promoting the malignant progression of colorectal cancer (Fig. 3a, b).
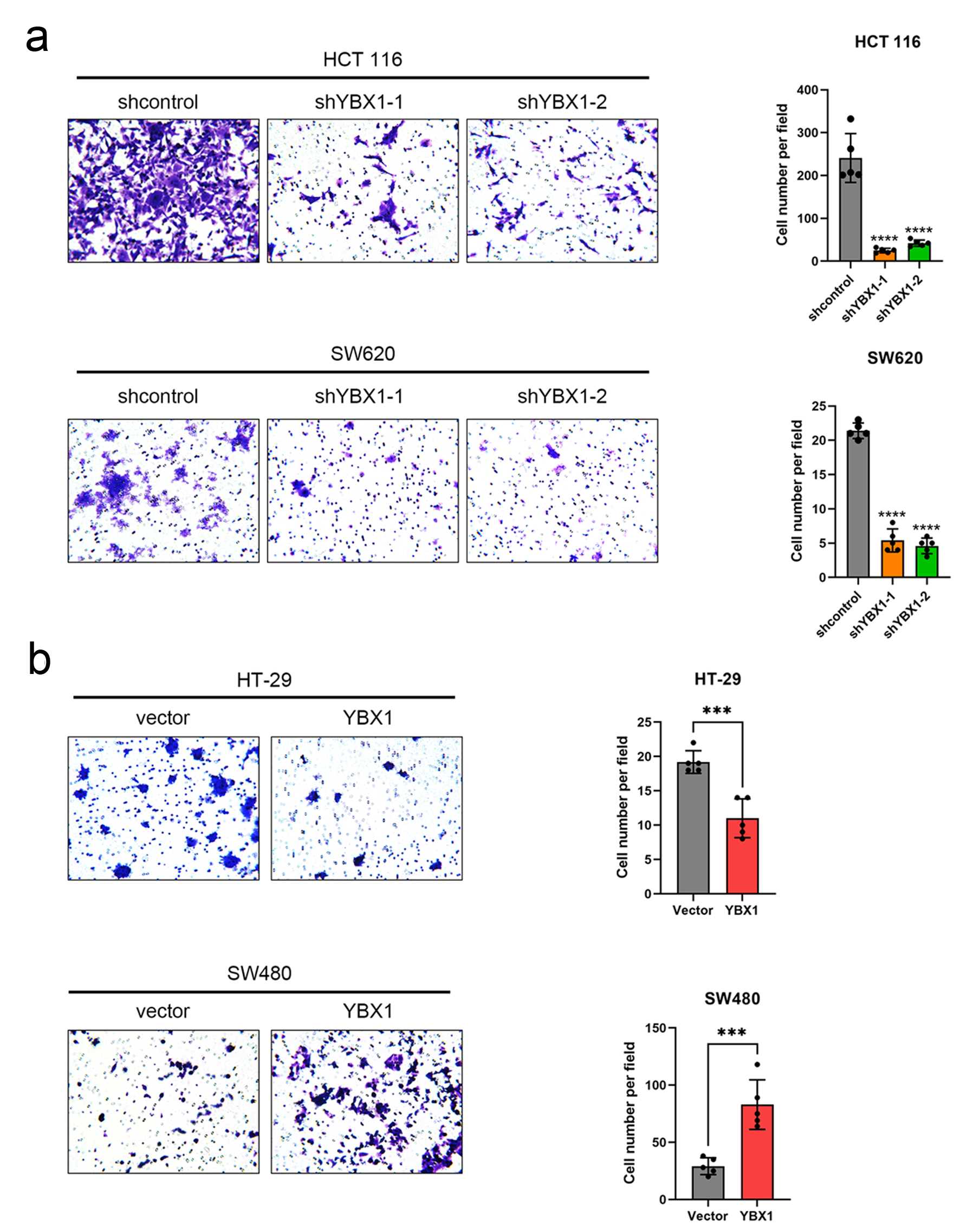 Click for large image | Figure 3. YBX1 promotes CRC cell migration. (a) Transwell migration assay showing that YBX1 knockdown significantly reduces migration ability in HCT 116 and SW620 cells. (b) YBX1 overexpression significantly enhances migration ability in HT-29 and SW480 cells. CRC: colorectal cancer; YBX1: Y-box binding protein 1. |
YBX1 functional differences in different cellular contexts may be related to its phosphorylation status
Overexpression of YBX1 in HT-29 cells may exhibit functional reversal due to multi-site phosphorylation, changing its structural state and resulting in a “dominant negative” phenotype. These findings underscore the importance of considering not only the total protein levels of YBX1 in targeted studies, but also its phosphorylation status and specific post-translational modification sites, which may critically influence its oncogenic potential.
YBX1 regulates HIF-1α expression and participates in hypoxic response
GSEA revealed that the HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY was significantly enriched in YBX1 high-expression samples in multiple GEO CRC cohorts (GSE21815, GSE31905, GSE35279, and GSE41657) (Fig. 4a).
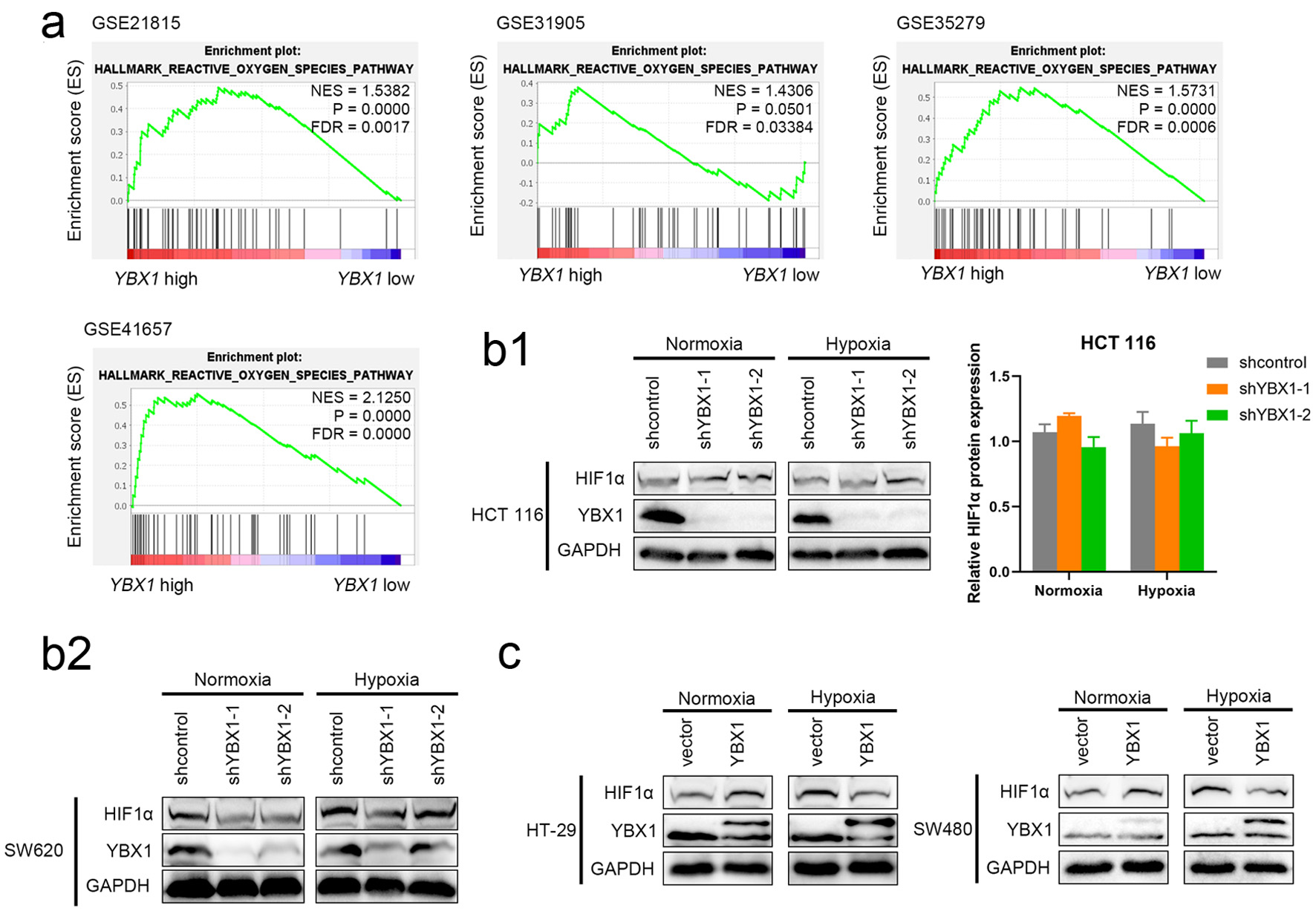 Click for large image | Figure 4. YBX1 regulates HIF-1α expression and participates in hypoxic response. (a) GSEA analysis showing significant enrichment of the ROS signaling pathway in YBX1 high-expression samples. (b) Effect of YBX1 knockdown on HIF-1α under normoxia and CoCl2-simulated hypoxia. (c) Effect of YBX1 overexpression on HIF-1α in HT-29 and SW480. GSEA: Gene Set Enrichment Analysis; HIF-1α: hypoxia-inducible factor 1-alpha; ROS: reactive oxygen species; YBX1: Y-box binding protein 1. |
To further explore the potential role of YBX1 in hypoxic responses, we established CRC cell models with YBX1 knockdown and overexpression, combined with CoCl2 treatment to simulate hypoxic conditions.
In SW620, YBX1 knockdown consistently reduced HIF-1α protein levels, and this reduction remained evident under hypoxia (Fig. 4b2). In HCT116, by contrast, YBX1 knockdown produced only a modest, non-significant change under normoxia and no robust difference under hypoxia (Fig. 4b1). In SW480 cells, YBX1 overexpression resulted in a significant increase in YBX1 protein levels under hypoxic conditions, but HIF-1α expression was lower than that in the control group (Fig. 4b, c).
Quantitative analysis confirmed these trends (Supplementary Material 1D-F, wjon.elmerpub.com).
RNA-seq suggests PI3K/AKT pathway may regulate YBX1 expression
To further explore the upstream regulatory mechanisms of YBX1 expression, we performed RNA sequencing analysis on YBX1 knockdown and control groups. The results revealed that multiple tumor-associated pathways, including PI3K/AKT and MAPK, were significantly downregulated upon YBX1 knockdown, suggesting its involvement in coordinating key oncogenic networks. Several downstream key genes showed decreased expression in the YBX1 knockdown group, suggesting that this pathway might serve as an important upstream regulator of YBX1 expression (Supplementary Materials 2 and 3, wjon.elmerpub.com).
PI3K inhibitor BKM120 downregulates YBX1 expression
To verify the regulatory relationship between the PI3K/AKT pathway and YBX1 expression, we treated CRC cells with the PI3K inhibitor BKM120 for 24 h. The results showed that BKM120 significantly inhibited the phosphorylation level of AKT (p-AKT expression decreased), while the total AKT expression remained unchanged. Meanwhile, YBX1 protein expression also significantly decreased, suggesting that YBX1 expression is positively regulated by the PI3K/AKT pathway (Fig. 5a-c). These findings reinforce the hypothesis that YBX1 is a downstream effector of PI3K/AKT signaling, and that its expression may be regulated through pathway-dependent phosphorylation events.
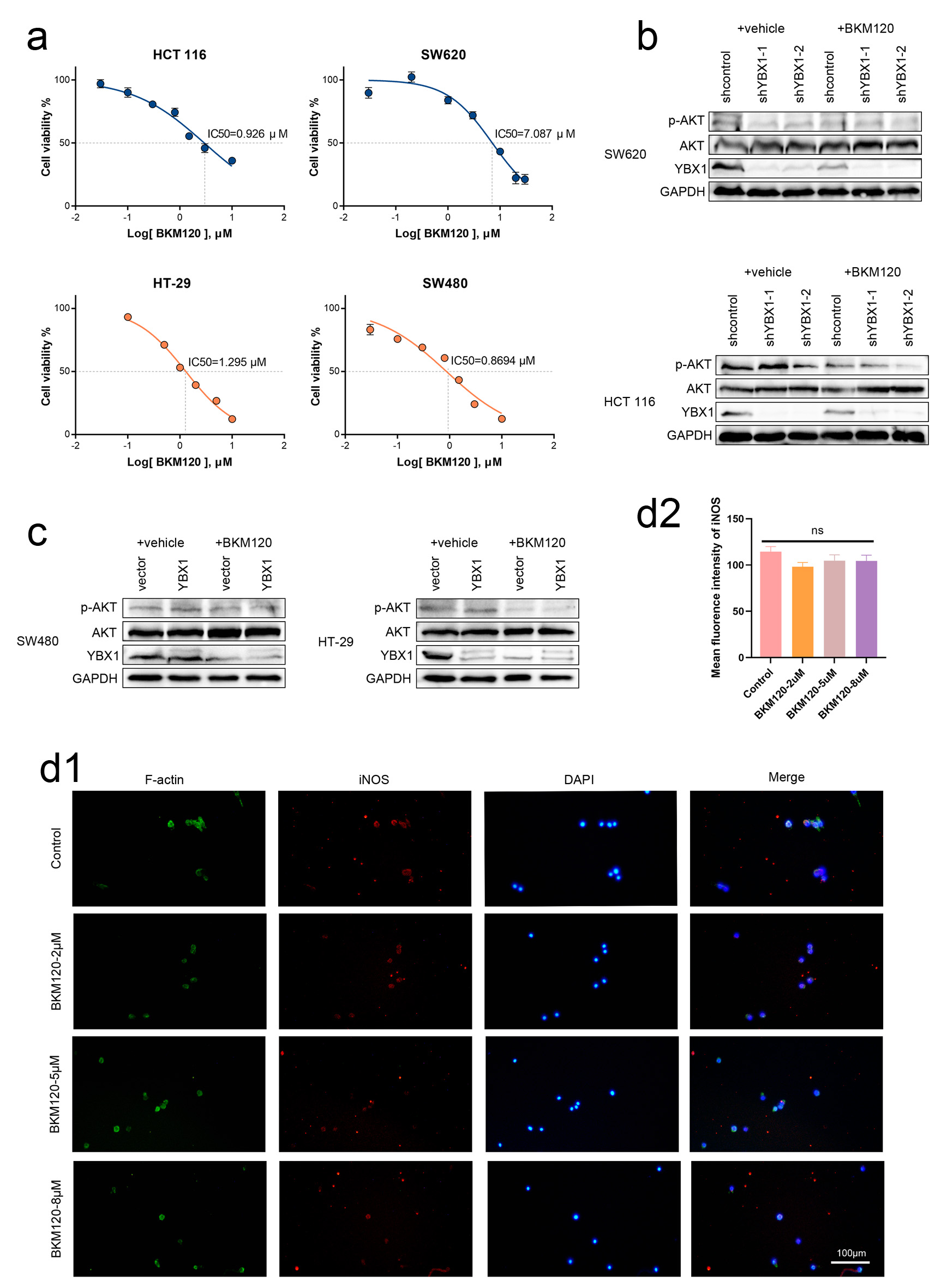 Click for large image | Figure 5. BKM120 downregulates YBX1 expression and promotes M1 polarization of macrophages. (a) IC50 values for various cell lines. (b, c) After 24-h BKM120 treatment of CRC cells, p-AKT and YBX1 protein levels significantly decreased, while total AKT levels did not change significantly. (d) Immunofluorescence staining showing that BKM120 treatment enhances iNOS expression in BMDMs. AKT: protein kinase B; BKM120: buparlisib; BMDMs: bone marrow-derived macrophages; CRC: colorectal cancer; IC50: half-maximal inhibitory concentration; iNOS: induced nitric oxide synthase; YBX1: Y-box binding protein 1. |
Inhibition of PI3K/AKT pathway affects macrophage polarization
To preliminarily explore the potential impact of PI3K/AKT/YBX1 pathway inhibition on the tumor-associated immune microenvironment, we further examined the effect of BKM120 on the polarization of BMDMs. BMDMs were exposed to different concentrations of BKM120 (0, 2, 5, and 8 µM) for 24 h, and immunofluorescence results showed that the expression of the M1 marker iNOS gradually increased, while the expression of the M2 marker Arg-1 decreased. These results indicate that PI3K/AKT signaling may influence macrophage polarization dynamics, and its inhibition promotes a pro-inflammatory M1 phenotype, which could enhance anti-tumor immunity in the CRC microenvironment (Figs. 5d and 6a).
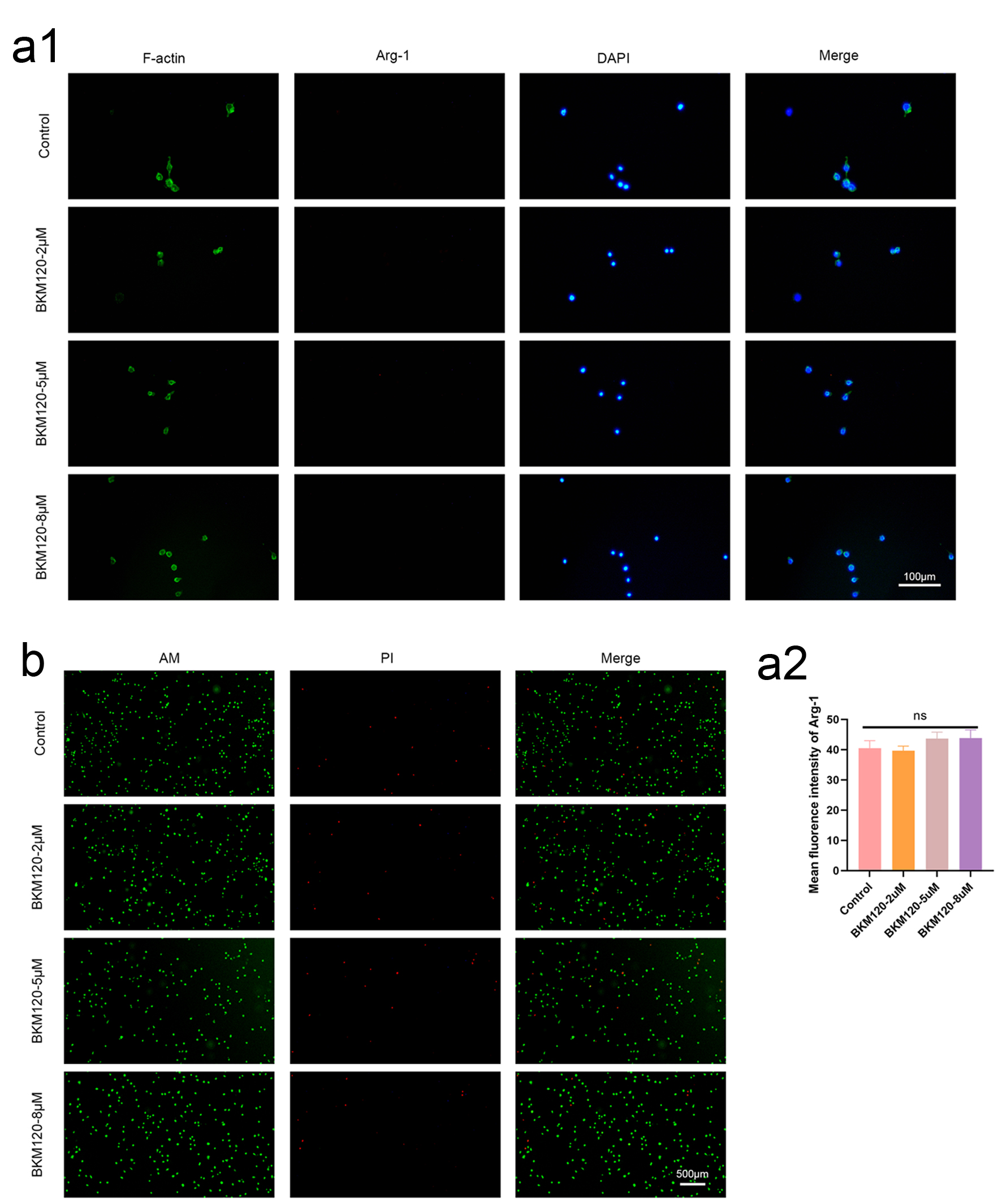 Click for large image | Figure 6. BKM120 promotes M1 polarization of macrophages and toxicity assessment. (a) Immunofluorescence staining showing decreased Arg-1 expression in BMDMs after BKM120 treatment. These results suggest that PI3K/AKT pathway inhibition might promote macrophage polarization towards M1, improving the tumor immune microenvironment. (b) The results show that at the concentrations of the drug used, L929 cells exhibited good vitality, indicating that the concentration of BKM120 is safe. AKT: protein kinase B; BKM120: buparlisib; BMDMs: bone marrow-derived macrophages; PI3K: phosphoinositide 3-kinase. |
Toxicity of BKM120 on L929 cells
To further evaluate the safety of BKM120 in CRC cell models, we conducted a toxicity experiment using L929 mouse fibroblasts. Cells were cultured with different concentrations of BKM120, and cell viability was assessed by live/dead staining (Calcein-AM/PI staining). The results showed that at the concentrations used, L929 cells exhibited good vitality (Fig. 6b). Staining results indicated that, under both normoxic and hypoxic conditions, the majority of the cells were green, indicating no cell death. These findings confirm that the tested concentrations of BKM120 exhibit minimal cytotoxicity in L929 fibroblasts, supporting its safety profile for further CRC-related studies.
| Discussion | ▴Top |
Increasing evidence suggests that in addition to genetic mutations, alterations in molecular regulatory networks play critical roles in driving CRC progression, therapy resistance, and immune evasion. Therefore, further elucidation of novel molecular mechanisms is essential for identifying new therapeutic strategies.
YBX1 is a multifunctional nucleic acid-binding protein, but the expression patterns, functional roles, and upstream regulation of YBX1 in CRC are not fully understood. In this study, we revealed that YBX1 is significantly overexpressed in CRC tissues and correlates with poor prognosis. Functional experiments further demonstrated that YBX1 promotes CRC cell proliferation and migration, and is positively regulated by PI3K/AKT signaling (Fig. 2).
The PI3K/AKT signaling pathway is a core pathway that regulates cell growth, metabolism, survival, and migration. Persistent activation of this pathway has been observed in various malignancies [12, 13]. Recent evidence suggests that PI3K/AKT signaling may promote tumor progression by modulating downstream effectors that drive phenotypic transformation and immune evasion in cancer cells [13]. However, it is unclear whether this pathway regulates CRC development via YBX1 activation [14].
Increasing attention has been paid to the role of YBX1 in the hypoxic tumor microenvironment. Recent studies have shown that YBX1 promotes HIF-1α expression via PI3K/AKT signaling, thereby enhancing tumor adaptation to hypoxia [15, 16]. Based on this, we hypothesized that YBX1 acts as a molecular bridge linking PI3K/AKT signaling and HIF-1α activation in CRC, thereby contributing to malignant progression (Fig. 7).
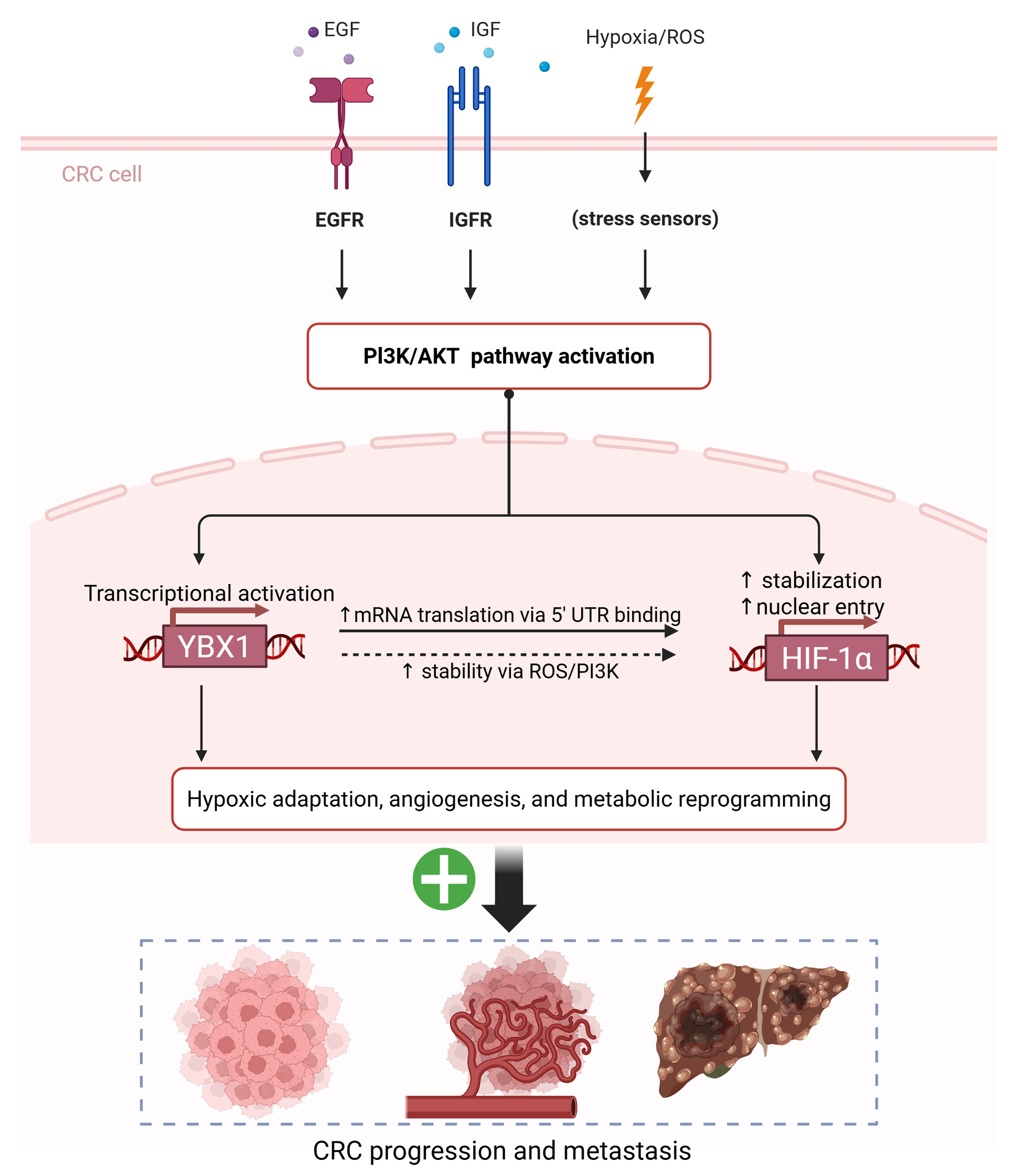 Click for large image | Figure 7. Proposed schematic model of the PI3K/AKT-YBX1-HIF-1α regulatory axis driving hypoxic adaptation and CRC progression. AKT: protein kinase B; CRC: colorectal cancer; HIF-1α: hypoxia-inducible factor 1-alpha; PI3K: phosphoinositide 3-kinase; YBX1: Y-box binding protein 1. |
RNA sequencing and GSEA analysis indicated that the PI3K/AKT signaling pathway might be involved in the upstream regulation of YBX1 expression. After treating CRC cells with the PI3K inhibitor BKM120, we observed a significant reduction in p-AKT and YBX1 protein levels, confirming the positive regulation of YBX1 by the PI3K/AKT pathway [17]. Notably, this concentration of BKM120 showed minimal cytotoxicity in L929 cells without significantly affecting cell viability (Fig. 6b), indicating its suitability for subsequent tumor microenvironment studies. Moreover, BKM120 treatment promoted M1 polarization of BMDMs, suggesting that the YBX1/PI3K/AKT axis may influence CRC progression by regulating the tumor immune microenvironment [18].
Notably, YBX1 overexpression in HT-29 cells suppressed proliferation and migration, contrasting with its typical oncogenic role in other CRC cell lines, suggesting a context-dependent regulatory mechanism. This phenomenon is different from its oncogenic effects in other cell lines. To address this discrepancy, we investigated the phosphorylation status of YBX1 and found that its overexpression in HT-29 cells was associated with significantly increased phosphorylation [19] (Fig. 2b). This observation implies that YBX1’s functional effects may be determined by cellular context and post-translational modifications, particularly site-specific phosphorylation.
Previous studies have demonstrated that YBX1 phosphorylation at different sites can lead to different outcomes. For instance, phosphorylation at Ser102 [20], which is mediated by kinases such as Akt, ribosomal S6 kinase (RSK), and mammalian target of rapamycin (mTOR), promotes the nuclear translocation and transcriptional activation of oncogenes such as epidermal growth factor receptor (EGFR) and Snail. This is consistent with YBX1’s established role as an oncogene [21, 22]. In contrast, phosphorylation at other sites (e.g., Ser165 [23], Thr80 [24], and Ser176 [19]) may impair nucleic acid binding, stability, or nuclear localization, potentially inducing transcriptional inactivation or loss of function [25] (Table 1). Notably, multi-site phosphorylation could result in a “dominant negative” or “inactive” conformation [26], explaining the tumor-suppressive phenotype observed in HT-29 cells (Figs. 2d and 3b).
 Click to view | Table 1. Impact of Different Phosphorylation Sites on YBX1 Functional Regulation |
Table 1 summarizes the kinase origins, functional directions, and regulatory mechanisms of various YBX1 phosphorylation sites reported in the current literature. The oncogenic or tumor-suppressive functions of YBX1 are finely regulated by these phosphorylation sites, with some sites enhancing its nuclear transcriptional regulatory ability, while others may lead to inactivation or mislocalization. Undiscovered sites offer potential directions for future mechanistic studies.
Previous studies have highlighted the dual roles of YBX1 phosphorylation in tumor biology depending on the cellular context. Bai et al [27] reported that PI3K/mTOR-mediated phosphorylation of YBX1, particularly at Ser102, promotes its nuclear translocation and the activation of proliferative transcriptional programs in basal-like head and neck cancer cells, thereby driving tumor progression. In contrast, in mesenchymal-like cells with low PI3K activity, reduced levels of phosphorylated YBX1 were associated with enhanced EMT programs and increased invasiveness, suggesting that the phosphorylation status of YBX1 critically influences tumor cell behavior. On the other hand, Sogorina et al [28] demonstrated that phosphorylation of YBX1 at Ser209 inhibits its nuclear translocation, thereby restraining its transcriptional activity. Taken together, these findings support the hypothesis that site-specific phosphorylation of YBX1 may exert opposite functional outcomes: phosphorylation at Ser102 enhances its oncogenic role, while phosphorylation at Ser209 may limit nuclear functions and confer a tumor-suppressive effect. This mechanistic framework may help to explain the differential effects that we observed in colorectal cancer cell lines and warrants further investigation into the precise contribution of individual phosphorylation sites to YBX1’s context-dependent roles in cancer progression.
The functional heterogeneity of YBX1 highlights the complexity of its regulation. While most CRC cell lines exhibit enhanced proliferation and migration when YBX1 is overexpressed, HT-29 cells display the opposite trend, which is likely due to differential phosphorylation patterns. These findings highlight the importance of cellular context and kinase activity in determining YBX1’s role, and suggest a potential mechanism for its dual functions in cancer progression. Further studies are needed to elucidate how specific phosphorylation signatures dictate YBX1’s switch between oncogenic and tumor-suppressive states [29].
Thus, we speculate that overexpression of YBX1 in HT-29 cells may result in functional reversal due to high-level phosphorylation, which changes its structural state. This observation suggests that in studies targeting YBX1, attention should not only be paid to its total protein levels, but also to its phosphorylation status and specific modification sites.
In addition to its role in regulating proliferation and migration through phosphorylation modifications, we also explored the involvement of YBX1 in hypoxic responses through the PI3K/AKT-HIF-1α axis. GSEA revealed that the HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY was significantly enriched in YBX1 high-expression samples in multiple GEO CRC cohorts (GSE21815, GSE31905, GSE35279, and GSE41657) (Fig. 4a). This pathway is closely associated with responses to oxidative stress and hypoxia responses, suggesting a potential role for YBX1 in regulating ROS-mediated stress pathways and hypoxic adaptation.
To further investigate this possibility, we established CRC cell models with YBX1 knockdown and overexpression, combined with CoCl2 treatment to simulate hypoxic conditions. Consistent with a cell line- and context-dependent relationship, YBX1 knockdown in SW620 reproducibly reduced HIF-1α under both normoxia and hypoxia, whereas in HCT116 the change was modest under normoxia and not robust under hypoxia (Fig. 4b; supported by densitometric quantification). In both SW480 and HT-29 cells, overexpression of YBX1 under hypoxic conditions led to a reduction in HIF-1α protein levels compared to vector control. This was particularly evident in SW480 cells (Fig. 4c), suggesting a possible negative feedback regulation mechanism mediated by YBX1 in response to hypoxia. Together, these findings indicate that YBX1 can either enhance or restrain HIF-1α in CRC cells depending on cellular background and stress intensity, highlighting its dual regulatory potential.
This phenomenon may be attributed to YBX1-mediated modulation of oxidative stress under hypoxic conditions, either through direct interaction with HIF-1α or through increased ROS accumulation, leading to the suppression of HIF-1α transcriptional activity [30]. Consistent with previous studies, YBX1 has been implicated in regulating tumor survival, angiogenesis, and invasiveness through oxidative stress pathways [29, 31, 32]. Our findings validated this regulatory trend across multiple CRC cell models, indicating that YBX1 acts as an upstream regulator of HIF-1α and participates in hypoxic stress responses.
Although YBX1 is generally considered to promote HIF-1α expression, our results demonstrate that under specific hypoxic conditions, YBX1 may inhibit excessive accumulation of HIF-1α through a negative feedback mechanism. These findings highlight the complex and context-dependent regulatory roles of YBX1 in tumor hypoxic microenvironments.
Moreover, considering that PI3K/AKT signaling is a well-known regulator of both hypoxic adaptation and HIF-1α expression, and that our previous experiments demonstrated PI3K/AKT-dependent regulation of YBX1 in CRC cells, it is reasonable to propose that PI3K/AKT activation promotes YBX1 upregulation, which in turn modulates HIF-1α dynamics under hypoxic conditions.
Based on our findings, we propose a conceptual model (Fig. 7) illustrating how the PI3K/AKT-YBX1-HIF-1α axis mediates hypoxia adaptation and promotes CRC progression and metastasis. In this model, PI3K/AKT signaling activates YBX1, which in turn enhances HIF-1α expression through translational and post-transcriptional mechanisms, especially under hypoxic and oxidative stress. This axis further drives angiogenesis and metabolic reprogramming, contributing to tumor adaptation.
Beyond its role in hypoxia, YBX1 may also influence immune modulation [33-35]. Future studies should explore whether YBX1 regulates the functional states of tumor-associated macrophages (TAMs), regulatory T cells (Tregs), and MDSCs [36, 37], thereby facilitating in immune evasion and providing new opportunities for immunotherapy.
Although our study demonstrated that BKM120 treatment promotes M1 polarization of BMDMs, suggesting its potential role in modulating the tumor immune microenvironment, the specific mechanisms by which BKM120 influences immune cell subsets such as TAMs, Tregs, and MDSCs in CRC remain to be elucidated. Future studies should investigate whether BKM120, alone or in combination with immunotherapies, can effectively modulate these immune cells to enhance anti-tumor immunity in CRC.
| Supplementary Material | ▴Top |
Suppl 1. Effect of YBX1 expression on mRNA levels in CRC cells under normoxic and hypoxic conditions.
Suppl 2. Heatmap of RNA-seq analysis.
Suppl 3. Heatmap and volcano plot of RNA-seq analysis.
Acknowledgments
The authors would like to thank Zhenjiang First People’s Hospital for their valuable support and assistance during this study.
Financial Disclosure
This research was funded by “Liu Ge Yi Gong Cheng” of Jiangsu Province (grant no. LGY2017022), Social Development Foundation of Zhenjiang (SH2022055), Medical Research Project of Yangzhou Health Commission 2023 (2023-2-28), and Science Foundation of the Affiliated People’s Hospital of Jiangsu University (Y2019021-S).
Conflict of Interest
The authors declare no conflict of interest.
Informed Consent
Not applicable.
Author Contributions
Hui Shan: conceptualization, validation, writing - original draft, writing - review and editing; Yan Wang: conceptualization, writing - review and editing; Siyu Hu: data curation and visualization; Yuting Wang: formal analysis and software; Rong Qin: funding acquisition; Niu Zhang: investigation; Guangyu Tian: methodology, resources, and supervision; Zhiyuan Qiu: project administration, supervision, writing - review and editing.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
Abbreviations
AKT: protein kinase B; BKM120: buparlisib (PI3K inhibitor); BMDMs: bone marrow-derived macrophages; CCK-8: cell counting kit-8; CKI: casein kinase I; CRC: colorectal cancer; EGFR: epidermal growth factor receptor; GEO: Gene Expression Omnibus; GEPIA: Gene Expression Profiling Interactive Analysis; GSEA: Gene Set Enrichment Analysis; IARC: International Agency for Research on Cancer; MDSCs: myeloid-derived suppressor cells; PBS: phosphate-buffered saline; PI3K: phosphoinositide 3-kinase; qRT-PCR: quantitative real-time polymerase chain reaction; ROS: reactive oxygen species; Ser: serine; TCGA: The Cancer Genome Atlas; Thr: threonine; Tregs: regulatory T cells; YBX1: Y-box binding protein 1
| References | ▴Top |
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424.
doi pubmed - Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145-164.
doi pubmed - Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249.
doi pubmed - Li H, Zhang D, Fu Q, Wang S, Wang Z, Zhang X, Chen X, et al. YBX1 as an oncogenic factor in T-cell acute lymphoblastic leukemia. Blood Adv. 2023;7(17):4874-4885.
doi pubmed - Xie Q, Zhao S, Liu W, Cui Y, Li F, Li Z, Guo T, et al. YBX1 Enhances Metastasis and Stemness by Transcriptionally Regulating MUC1 in Lung Adenocarcinoma. Front Oncol. 2021;11:702491.
doi pubmed - Krishnamoorthy HR, Karuppasamy R. Deciphering the prognostic landscape of triple-negative breast cancer: A focus on immune-related hub genes and therapeutic implications. Biotechnol Appl Biochem. 2025;72(3):825-845.
doi pubmed - Tang T, Yang T, Xue H, Liu X, Yu J, Liang C, Li D, et al. Breast cancer stem cell-derived exosomal lnc-PDGFD induces fibroblast-niche formation and promotes lung metastasis. Oncogene. 2025;44(9):601-617.
doi pubmed - Zhuo H, Hou J, Hong Z, Yu S, Peng H, Zhang L, Xie W, et al. TAGLN2 induces resistance signature ISGs by activating AKT-YBX1 signal with dual pathways and mediates the IFN-related DNA damage resistance in gastric cancer. Cell Death Dis. 2024;15(8):608.
doi pubmed - Huang M, Sun J, Jiang Q, Zhao X, Huang H, Lei M, Jiang S, et al. CircKIAA0182-YBX1 axis: a key driver of lung cancer progression and chemoresistance. Cancer Lett. 2025;612:217494.
doi pubmed - Li Z, Lu W, Yin F, Huang A. YBX1 as a prognostic biomarker and potential therapeutic target in hepatocellular carcinoma: a comprehensive investigation through bioinformatics analysis and in vitro study. Transl Oncol. 2024;45:101965.
doi pubmed - Hou P, Chen F, Yong H, Lin T, Li J, Pan Y, Jiang T, et al. PTBP3 contributes to colorectal cancer growth and metastasis via translational activation of HIF-1alpha. J Exp Clin Cancer Res. 2019;38(1):301.
doi pubmed - Leiphrakpam PD, Are C. PI3K/Akt/mTOR signaling pathway as a target for colorectal cancer treatment. Int J Mol Sci. 2024;25(6).
doi pubmed - Maharati A, Moghbeli M. PI3K/AKT signaling pathway as a critical regulator of epithelial-mesenchymal transition in colorectal tumor cells. Cell Commun Signal. 2023;21(1):201.
doi pubmed - Sheng Z, Luo S, Huang L, Deng YN, Zhang N, Luo Y, Zhao X, et al. SENP1-mediated deSUMOylation of YBX1 promotes colorectal cancer development through the SENP1-YBX1-AKT signaling axis. Oncogene. 2025;44(19):1361-1374.
doi pubmed - Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721-732.
doi pubmed - Zhao J, Zhang P, Wang X. YBX1 promotes tumor progression via the PI3K/AKT signaling pathway in laryngeal squamous cell carcinoma. Transl Cancer Res. 2021;10(11):4859-4869.
doi pubmed - Liu T, Xie XL, Zhou X, Chen SX, Wang YJ, Shi LP, Chen SJ, et al. Y-box binding protein 1 augments sorafenib resistance via the PI3K/Akt signaling pathway in hepatocellular carcinoma. World J Gastroenterol. 2021;27(28):4667-4686.
doi pubmed - Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol. 2017;198(3):1006-1014.
doi pubmed - Martin M, Hua L, Wang B, Wei H, Prabhu L, Hartley AV, Jiang G, et al. Novel serine 176 phosphorylation of YBX1 activates NF-kappaB in colon cancer. J Biol Chem. 2017;292(8):3433-3444.
doi pubmed - Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, et al. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol. 2006;26(1):277-292.
doi pubmed - Jiang D, Qiu T, Peng J, Li S, Tala, Ren W, Yang C, et al. YB-1 is a positive regulator of KLF5 transcription factor in basal-like breast cancer. Cell Death Differ. 2022;29(6):1283-1295.
doi pubmed - Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24(26):4281-4292.
doi pubmed - Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15(5):402-415.
doi pubmed - Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5(1):95-110.
doi pubmed - Kretov DA, Mordovkina DA, Eliseeva IA, Lyabin DN, Polyakov DN, Joshi V, Desforges B, et al. Inhibition of transcription induces phosphorylation of YB-1 at Ser102 and its accumulation in the nucleus. Cells. 2019;9(1).
doi pubmed - Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc). 2011;76(13):1402-1433.
doi pubmed - Bai Y, Gotz C, Chincarini G, Zhao Z, Slaney C, Boath J, Furic L, et al. YBX1 integration of oncogenic PI3K/mTOR signalling regulates the fitness of malignant epithelial cells. Nat Commun. 2023;14(1):1591.
doi pubmed - Sogorina EM, Kim ER, Sorokin AV, Lyabin DN, Ovchinnikov LP, Mordovkina DA, Eliseeva IA. YB-1 phosphorylation at serine 209 inhibits its nuclear translocation. Int J Mol Sci. 2021;23(1):428.
doi pubmed - El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, et al. Translational activation of HIF1alpha by YB-1 promotes sarcoma metastasis. Cancer Cell. 2015;27(5):682-697.
doi pubmed - Liu C, Du H, Yu G, Qi J, Dong H, Hu R, Wang F, et al. Chronic stress stimulates protumor macrophage polarization to propel lung cancer progression. Cancer Res. 2025;85(13):2429-2447.
doi pubmed - Wu F, Li D. YB1 and its role in osteosarcoma: a review. Front Oncol. 2024;14:1452661.
doi pubmed - Chen SP, Zhu GQ, Xing XX, Wan JL, Cai JL, Du JX, Song LN, et al. LncRNA USP2-AS1 Promotes Hepatocellular Carcinoma Growth by Enhancing YBX1-Mediated HIF1alpha Protein Translation Under Hypoxia. Front Oncol. 2022;12:882372.
doi pubmed - Cui R, Luo Z, Zhang X, Yu X, Yuan G, Li X, Xie F, et al. Targeting PI3K signaling to overcome tumor immunosuppression: synergistic strategies to enhance cancer vaccine efficacy. Vaccines (Basel). 2025;13(3).
doi pubmed - Okkenhaug K, Graupera M, Vanhaesebroeck B. Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 2016;6(10):1090-1105.
doi pubmed - Sai J, Owens P, Novitskiy SV, Hawkins OE, Vilgelm AE, Yang J, Sobolik T, et al. PI3K inhibition reduces mammary tumor growth and facilitates antitumor immunity and anti-PD1 responses. Clin Cancer Res. 2017;23(13):3371-3384.
doi pubmed - Li Y, Jin H, Li Q, Shi L, Mao Y, Zhao L. The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol Cancer. 2024;23(1):130.
doi pubmed - Xiang J, Wang J, Xiao H, Huang C, Wu C, Zhang L, Qian C, et al. Targeting tumor-associated macrophages in colon cancer: mechanisms and therapeutic strategies. Front Immunol. 2025;16:1573917.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.