Figures
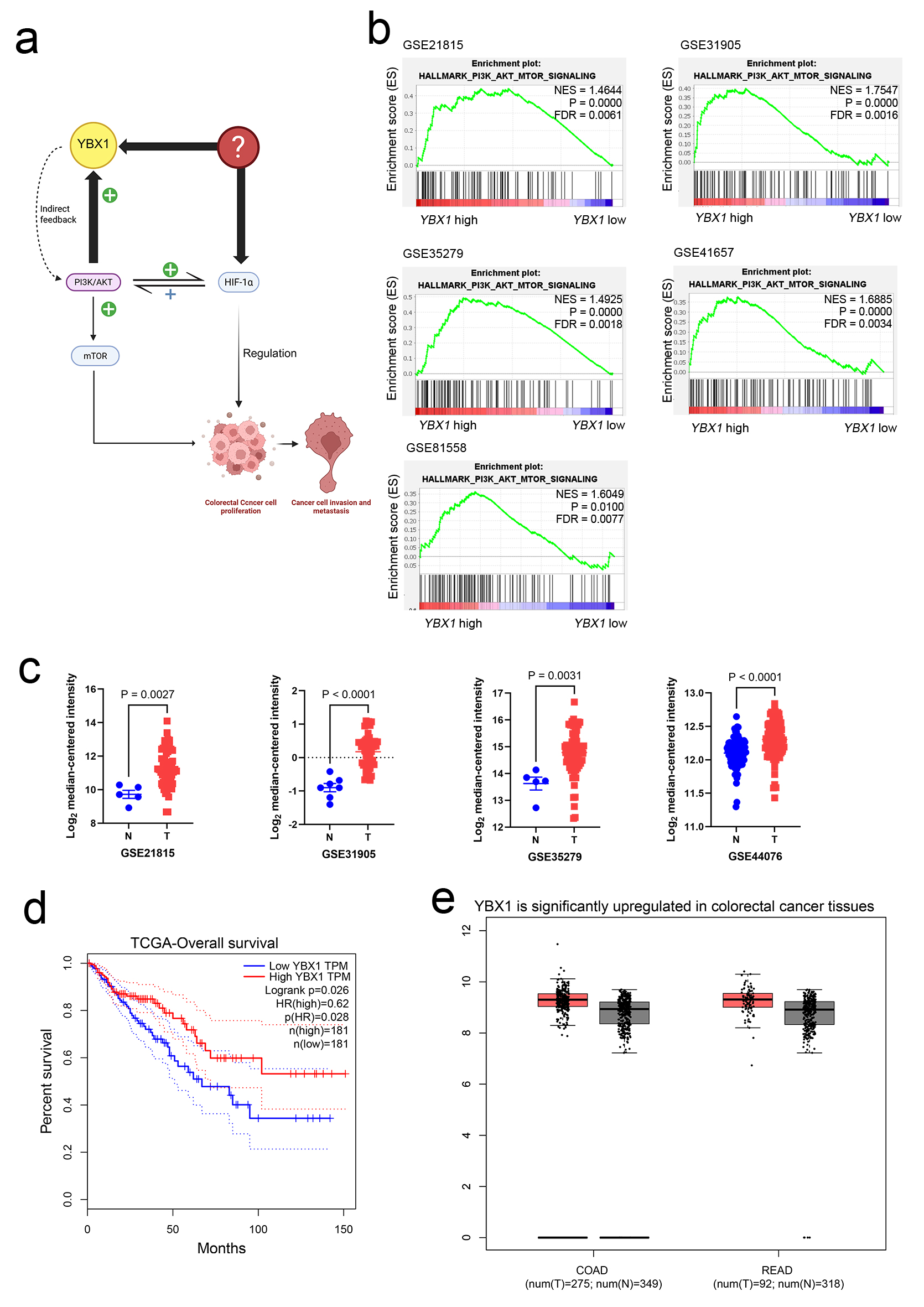
Figure 1. YBX1 high expression in CRC and its association with the PI3K/AKT pathway. (a) Schematic diagram showing the potential mechanism of YBX1 in CRC. (b) GSEA showing significant enrichment of the PI3K/AKT/mTOR signaling pathway in YBX1 high-expression samples. (c) GEO database analysis (GSE21815, GSE31905, GSE35279, and GSE44076) showing significant upregulation of YBX1 expression in COAD and READ tissues. (d) TCGA database analysis showing that high YBX1 expression is associated with poor survival prognosis in CRC patients. (e) TCGA data further validating the high expression of YBX1 in COAD and READ tumor tissues. AKT: protein kinase B; COAD: colon adenocarcinoma; CRC: colorectal cancer; GEO: Gene Expression Omnibus; GSEA: Gene Set Enrichment Analysis; mTOR: mammalian target of rapamycin; PI3K: phosphoinositide 3-kinase; READ: rectum adenocarcinoma; TCGA: The Cancer Genome Atlas; YBX1: Y-box binding protein 1.
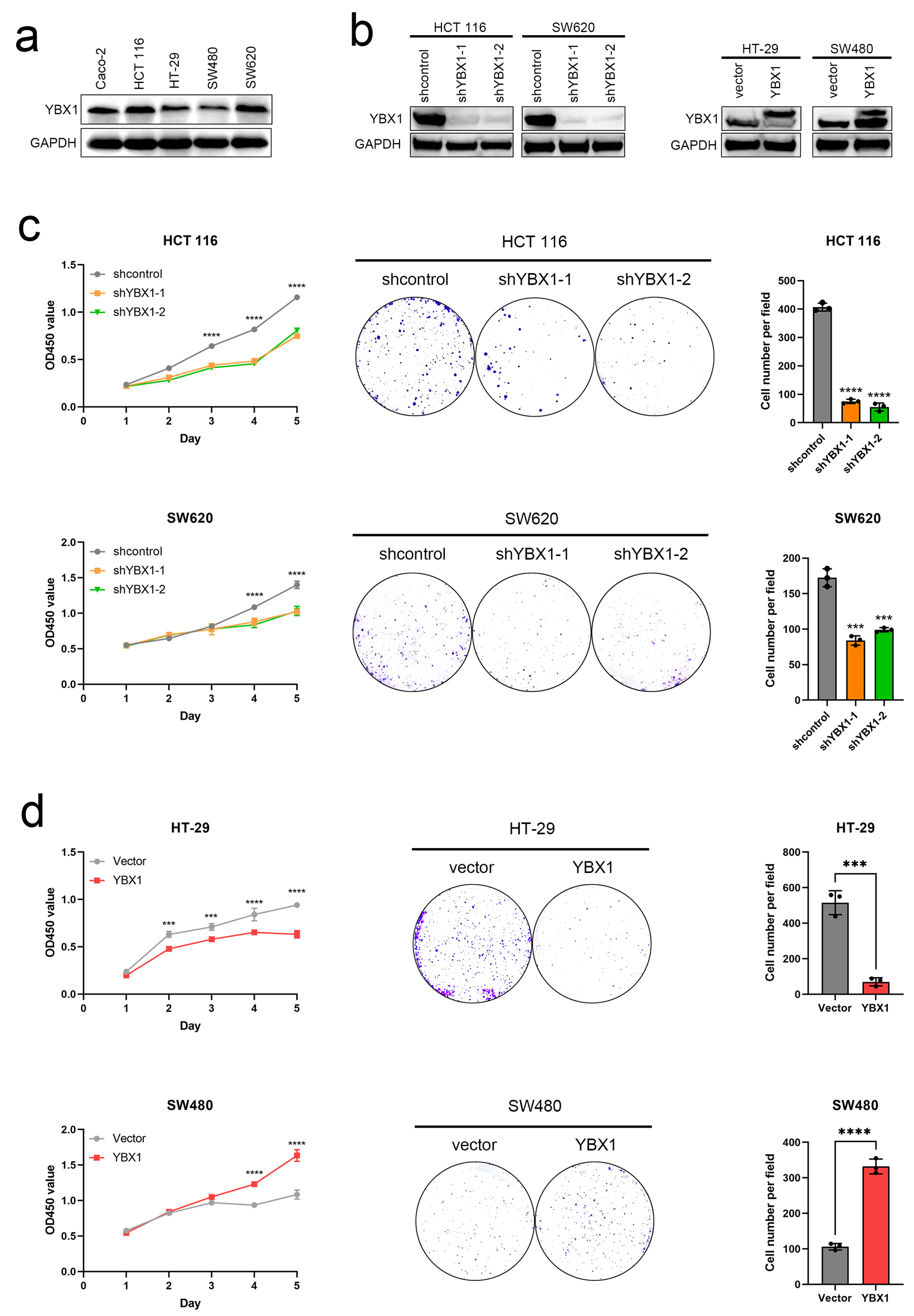
Figure 2. YBX1 regulates CRC cell proliferation and colony formation. (a) Expression levels of YBX1 in five CRC cell lines. (b) Western blot verification of YBX1 knockdown in HCT 116 and SW620 cells and YBX1 overexpression in HT-29 and SW480 cells. (c) CCK-8 assays showing that YBX1 knockdown inhibits cell proliferation, while overexpression promotes cell proliferation. (d) Colony formation assays showing that YBX1 knockdown decreases colony formation ability, while overexpression enhances colony formation ability. CCK-8: cell counting kit-8; CRC: colorectal cancer; YBX1: Y-box binding protein 1.
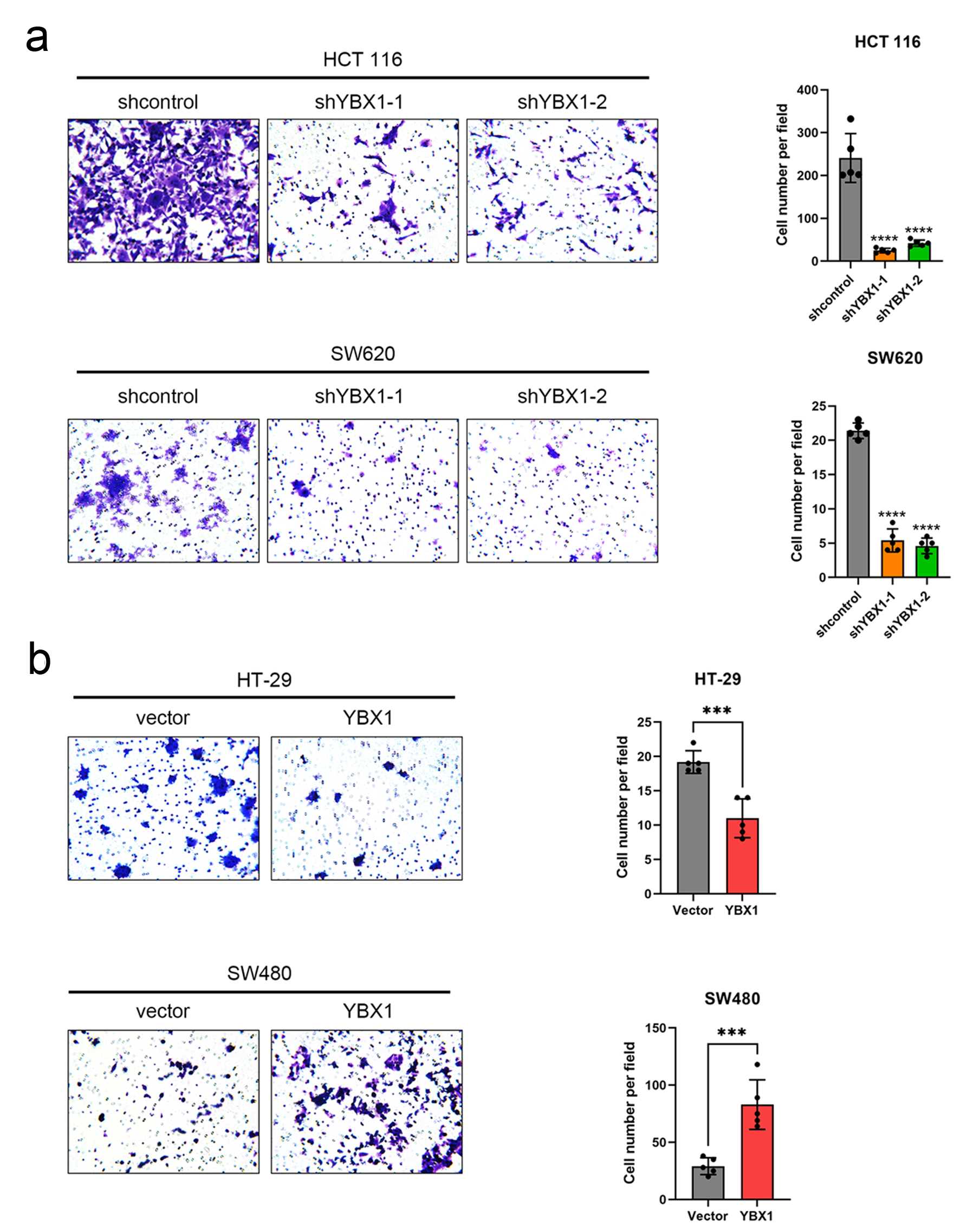
Figure 3. YBX1 promotes CRC cell migration. (a) Transwell migration assay showing that YBX1 knockdown significantly reduces migration ability in HCT 116 and SW620 cells. (b) YBX1 overexpression significantly enhances migration ability in HT-29 and SW480 cells. CRC: colorectal cancer; YBX1: Y-box binding protein 1.
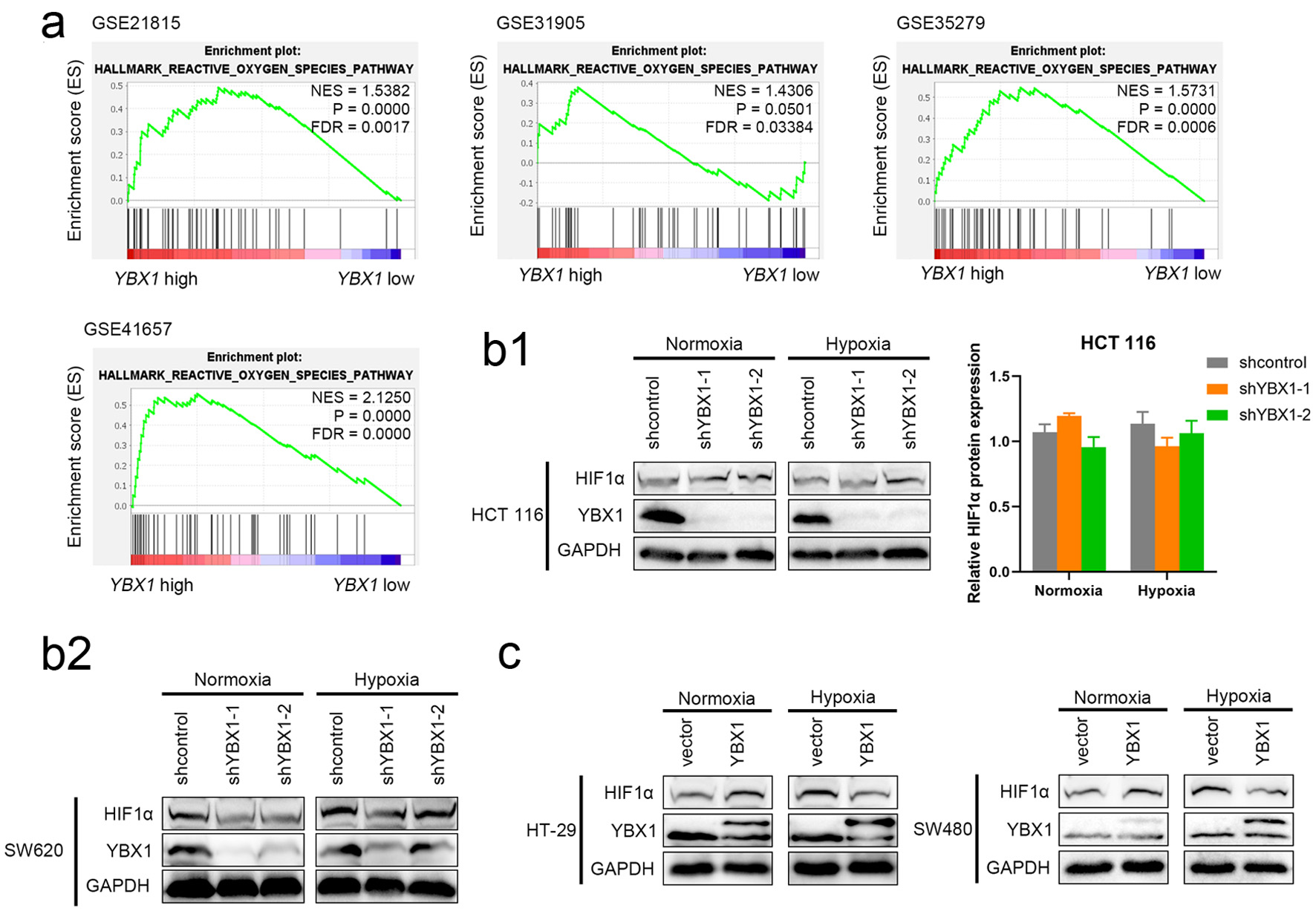
Figure 4. YBX1 regulates HIF-1α expression and participates in hypoxic response. (a) GSEA analysis showing significant enrichment of the ROS signaling pathway in YBX1 high-expression samples. (b) Effect of YBX1 knockdown on HIF-1α under normoxia and CoCl2-simulated hypoxia. (c) Effect of YBX1 overexpression on HIF-1α in HT-29 and SW480. GSEA: Gene Set Enrichment Analysis; HIF-1α: hypoxia-inducible factor 1-alpha; ROS: reactive oxygen species; YBX1: Y-box binding protein 1.
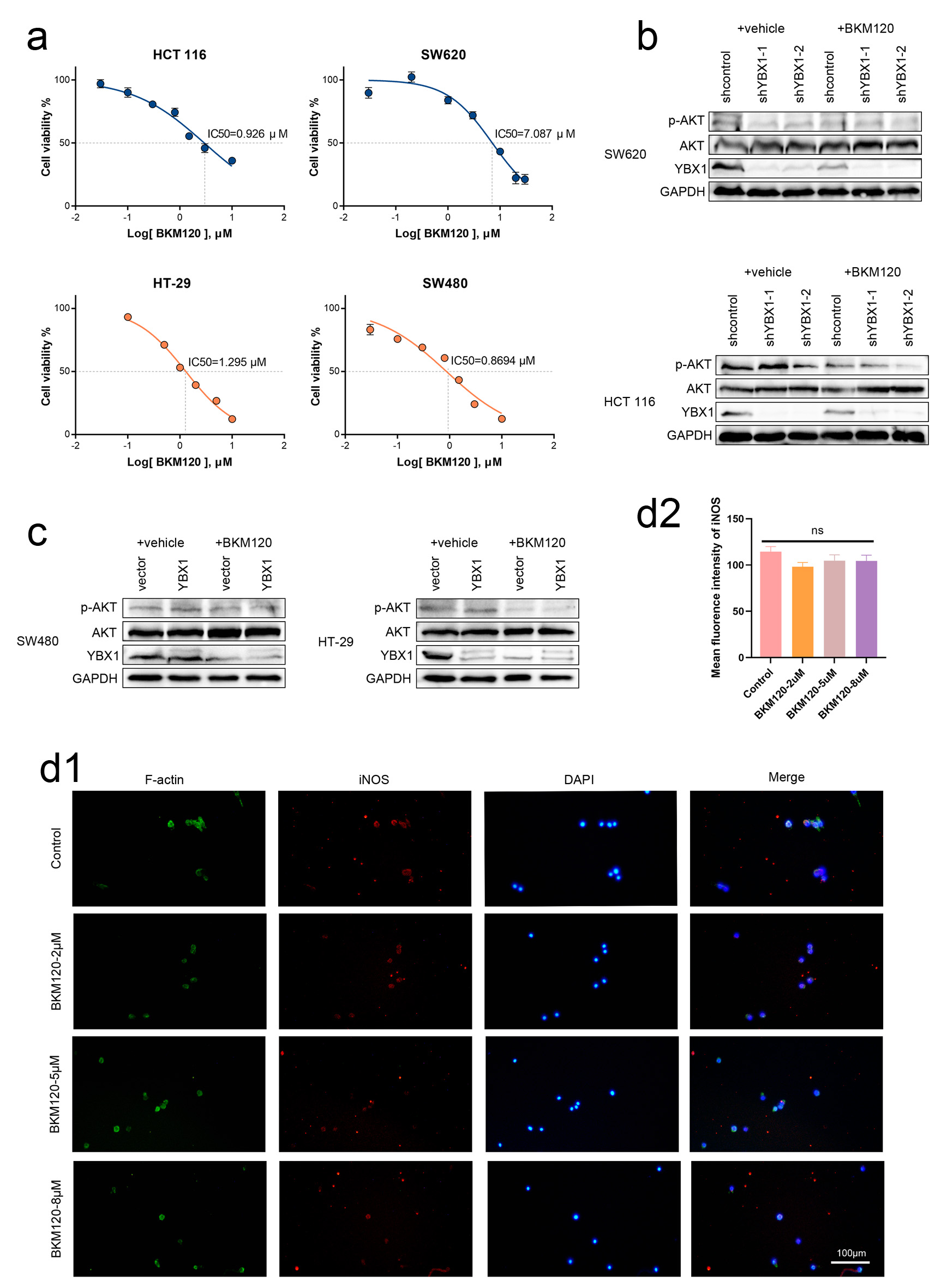
Figure 5. BKM120 downregulates YBX1 expression and promotes M1 polarization of macrophages. (a) IC50 values for various cell lines. (b, c) After 24-h BKM120 treatment of CRC cells, p-AKT and YBX1 protein levels significantly decreased, while total AKT levels did not change significantly. (d) Immunofluorescence staining showing that BKM120 treatment enhances iNOS expression in BMDMs. AKT: protein kinase B; BKM120: buparlisib; BMDMs: bone marrow-derived macrophages; CRC: colorectal cancer; IC50: half-maximal inhibitory concentration; iNOS: induced nitric oxide synthase; YBX1: Y-box binding protein 1.
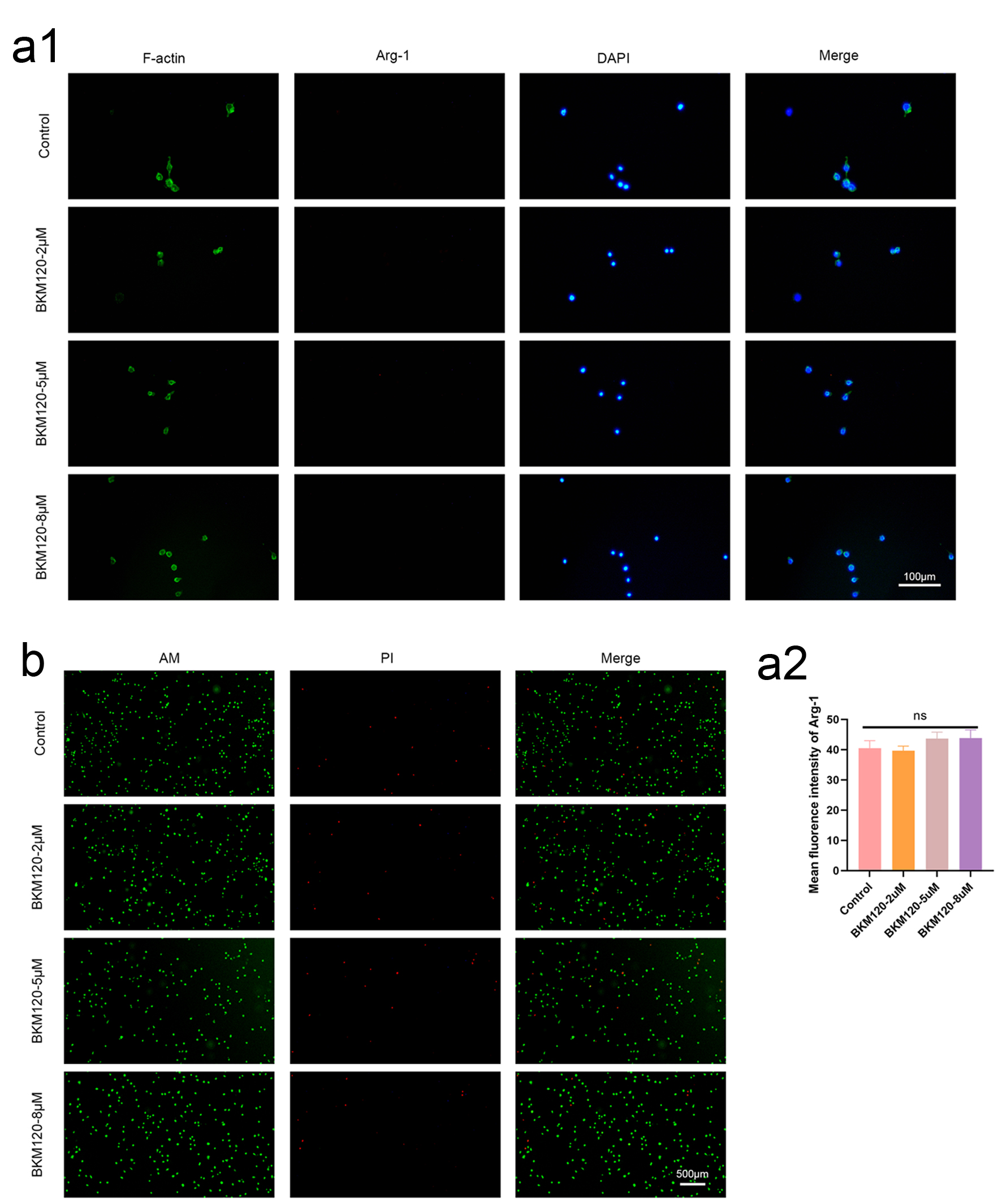
Figure 6. BKM120 promotes M1 polarization of macrophages and toxicity assessment. (a) Immunofluorescence staining showing decreased Arg-1 expression in BMDMs after BKM120 treatment. These results suggest that PI3K/AKT pathway inhibition might promote macrophage polarization towards M1, improving the tumor immune microenvironment. (b) The results show that at the concentrations of the drug used, L929 cells exhibited good vitality, indicating that the concentration of BKM120 is safe. AKT: protein kinase B; BKM120: buparlisib; BMDMs: bone marrow-derived macrophages; PI3K: phosphoinositide 3-kinase.
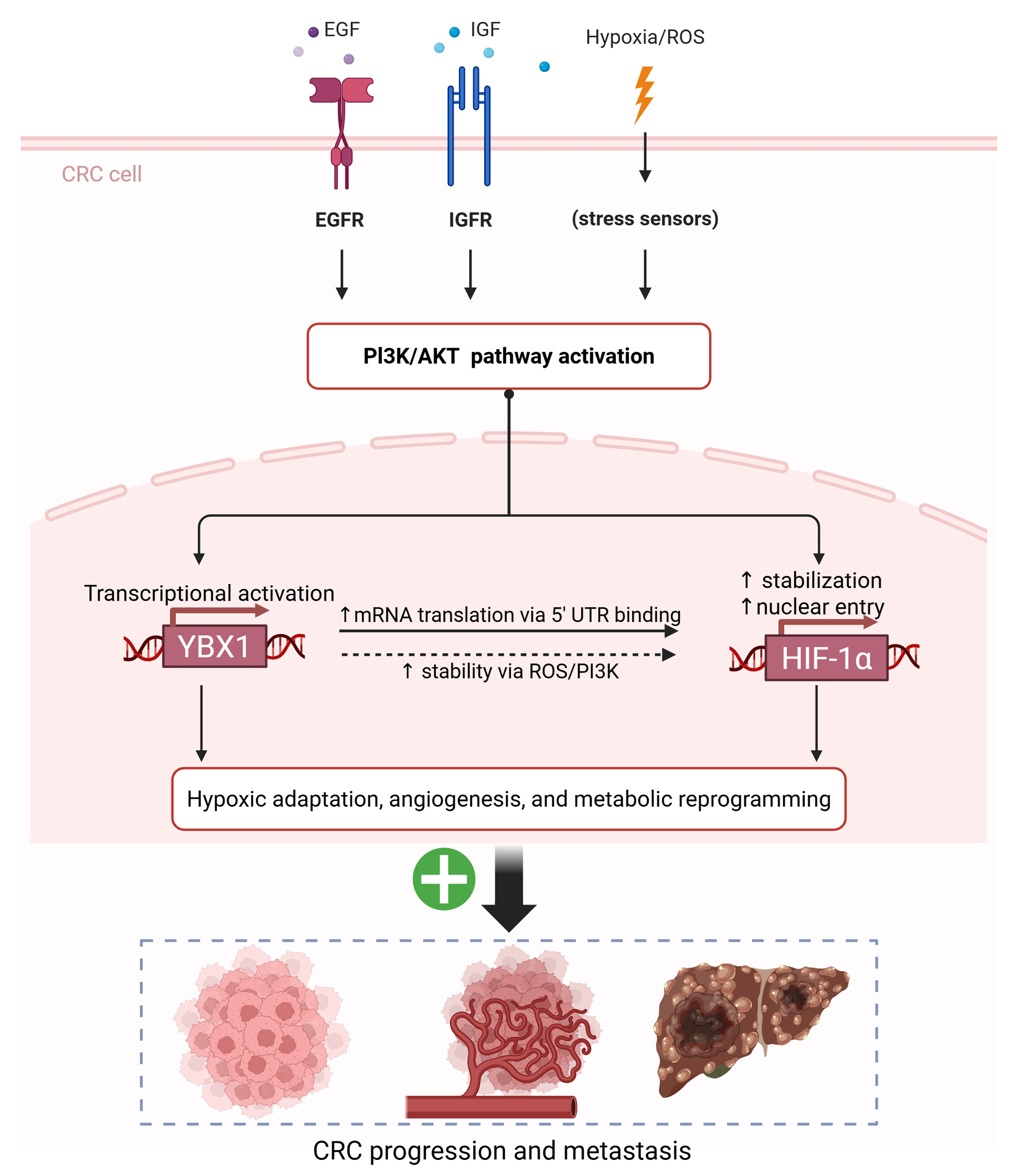
Figure 7. Proposed schematic model of the PI3K/AKT-YBX1-HIF-1α regulatory axis driving hypoxic adaptation and CRC progression. AKT: protein kinase B; CRC: colorectal cancer; HIF-1α: hypoxia-inducible factor 1-alpha; PI3K: phosphoinositide 3-kinase; YBX1: Y-box binding protein 1.
Table
Table 1. Impact of Different Phosphorylation Sites on YBX1 Functional Regulation
| Phosphorylation site | Regulating kinase | Functional direction | Mechanism description |
|---|
| AKT: protein kinase B; CKI: casein kinase I; CRC: colorectal cancer; EGFR: epidermal growth factor receptor; IL: interleukin; mTOR: mammalian target of rapamycin; NF-κB: nuclear factor-κB; RSK: ribosomal S6 kinase; Ser: serine; Thr: threonine; YBX1: Y-box binding protein 1. |
| Ser102 [20, 22] | Akt, RSK, mTOR | Oncogenic | Promotes YBX1 nuclear translocation, enhances its transcriptional activation ability, and regulates the expression of target genes such as EGFR and Snail. |
| Ser165 [23] | Unknown | Tumor suppressive (speculated) | May lead to inhibition of YBX1’s transcriptional activity; further verification is needed. |
| Ser176 [19] | CKI (indirect regulation, induced by IL-1β) | Oncogenic | Phosphorylation at Ser176 activates the NF-κB signaling pathway in CRC cells, promoting YBX1 nuclear translocation and transcriptional activation, an important regulatory point for its oncogenic function. |
| Thr80 [24] | Unknown | Unclear | May affect YBX1 stability and its RNA-binding ability. |
| Multi-site phosphorylation [26] | Multi-pathway activation | Biphasic function | Excessive phosphorylation may cause nuclear exclusion or non-functional aggregation, resulting in transcriptional inactivation or a “dominant negative” effect. |






