| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 000, Number 000, October 2025, pages 000-000
A Simplified Novel Algorithm to Predict the 21-Gene Recurrence Score
Maher A. Sughayera, e, Bayan Maraqaa, Batool Qura’anb, Ahmad Alsughayerc, Hikmat Abdel-Razeqd
aDepartment of Pathology and Laboratory Medicine, King Hussein Cancer Center, Amman, Jordan
bMinistry of Health, Amman, Jordan
cDepartment of Surgery, King Hussein Cancer Center, Amman, Jordan
dDepartment of Medical Oncology, King Hussein Cancer Center, Amman, Jordan
eCorresponding Author: Maher A. Sughayer, Department of Pathology and Laboratory Medicine, King Hussein Cancer Center, Amman 11941, Jordan
Manuscript submitted June 29, 2025, accepted August 14, 2025, published online October 10, 2025
Short title: Predicting the 21-Gene Recurrence Score
doi: https://doi.org/10.14740/wjon2634
| Abstract | ▴Top |
Background: The 21-gene recurrence score (Oncotype DX) guides adjuvant chemotherapy decisions in early-stage estrogen receptor (ER)-positive, human epidermal growth factor receptor-2 (HER2)-negative breast cancer. However, its high cost and limited availability motivate the development of simplified predictive models using routinely reported pathology parameters. The aim of this study was to develop and validate a practical, rule-based algorithm that predicts Oncotype DX recurrence score (RS) category using only histologic grade and progesterone receptor (PR) expression percentage.
Methods: This retrospective study included 528 patients with ER+/HER2- early breast cancer who underwent Oncotype DX testing. Cases were randomly assigned to a learning (n = 377) and validation (n = 151) set. Univariate analysis and receiver operating characteristics (ROC) curves were used to determine PR% cut-offs within each histologic grade to stratify patients into low-risk (RS ≤ 25) or high-risk (RS > 25) categories. A stepwise algorithm was derived from these parameters and tested in the validation cohort.
Results: Histologic grade and PR% were significantly associated with RS. Grade 1 tumors were uniformly low-risk regardless of PR%. In grade 2, PR ≥ 60% achieved 100% sensitivity for low RS; in grade 3, PR < 40% achieved 100% sensitivity for high risk. The algorithm confidently stratified ∼ 65% of cases. In the validation set, the model showed 87.5% sensitivity, 100% specificity, 100% positive predictive value (PPV), 99% negative predictive value (NPV), and 99% overall accuracy.
Conclusion: This simplified algorithm accurately predicts Oncotype DX RS category using only histologic grade and PR%. It enables confident risk stratification in most patients without molecular testing, offering a low-cost, practical tool for clinical decision-making, particularly in resource-limited settings.
Keywords: 21-gene recurrence score; Oncotype DX; Breast cancer; Predictive algorithm; Progesterone receptor; Histologic grade
| Introduction | ▴Top |
Breast cancer is the most common tumor among women globally and is the primary cause of cancer-related mortality in women, according to the 2022 updated estimates from the International Agency for Research on Cancer (IARC) [1].
Estrogen receptor (ER)-positive, human epidermal growth factor receptor-2 (HER2)-negative breast cancer constitutes approximately 70% of all breast cancer cases, with the majority of patients exhibiting a favorable response to endocrine therapy alone, with no benefit from adjuvant chemotherapy. However, a subset of patients harbor more aggressive disease that may benefit from the addition of chemotherapy [2].
To guide adjuvant treatment decisions in this group, the 21-gene recurrence score, commercially known as Oncotype DX (ODx) multigene expression assay is widely used for patients with early-stage, ER-positive, HER2-negative breast cancer. This assay generates a recurrence score (RS) ranging from 0 to 100. A RS of 25 or lower is classified as low risk, indicating limited benefit from chemotherapy, whereas a score of 26 or higher denotes high risk, for which adjuvant chemotherapy may offer clinical benefit [3]. Beyond its prognostic utility, ODx is the only assay currently validated as predictive of chemotherapy benefit and is endorsed as such by leading professional societies, including the American Society of Clinical Oncology (ASCO) [4] and the National Comprehensive Cancer Network (NCCN) [5]. It is also the sole multigene test incorporated into the pathological prognostic staging system and has been granted a “preferred” status within the NCCN clinical guidelines [5].
Despite its clinical value, ODx is expensive and has a turnaround time of approximately 2 weeks which can pose challenges in resource-limited settings. As a result, several studies have increasingly explored simpler, faster, and more affordable alternatives. Many of these studies have aimed to identify clinicopathological features that could either predict the actual ODx RS or at least estimate the likelihood of a high-risk result, using information that is already available in routine pathology reports [5-31].
Several of these alternatives use predictive equations or models derived by regression analysis and/or machine learning, based on routinely assessed pathological variables. These variables may be readily available in breast cancer pathology reports. However, some models require more details and/or processing to be usable in the calculations. For instance, the Magee equations incorporate the H-score for ER and progesterone receptor (PR) which may need to be calculated, in addition to the Nottingham score and Ki-67 proliferation index [25, 29]. In practice, most pathology reports may not include the H-score or the details of the Nottingham score for the grade or the Ki-67, which can limit the direct applicability of such models.
Other approaches, such as the nomogram developed at the University of Tennessee Medical Center, require very minimal basic information that is readily available in the pathology reports. These include tumor size, histologic grade (without the need for the full Nottingham score), PR status (rather than percentage or H-score), patient age, presence or absence of lymphovascular invasion (LVI), and histologic tumor type [30]. More recently, this nomogram has been refined using machine learning models to predict low-risk (0 - 25) or high-risk (26 - 100) ODx categories. The updated model uses either non-quantitative ER/PR or quantitative ER/PR with or without Ki-67, offering additional flexibility for clinical use [15].
The performance of these models in terms of prediction of a high- or low-risk ODx scores is generally good, and varies in C-index from 0.81 to 0.87 [15, 30]. For instance, the Magee algorithm can potentially identify approximately 70% of patients as low-risk without ODx testing, with an accuracy of 95.1% [27]. On the other hand, the proportion of those patients based on the University of Chicago model, using a high-sensitivity rule-out threshold, is up to 43% [15].
Many of the studies published so far have revealed that PR status or expression level, along with histologic grade, are the most significant predictors for ODx score, and the most important prognostic factors for breast cancer in general [8, 22, 30].
The objective of this study was to find a simple method using routinely assessed parameters that are readily available in the patients’ pathology reports (without the need for calculations or online nomograms) to predict the ODx risk category (low risk or high risk) in ER-positive, HER2-negative early breast cancer. Specifically, we aimed to evaluate the predictive utility of PR expression and histologic grade, and to estimate the proportion of patients for whom ODx testing could be safely omitted based on these parameters.
| Materials and Methods | ▴Top |
This retrospective study was approved by the Institutional Review Board (IRB) at King Hussein Cancer Center under approval number 24-KHCC-108. This study was conducted in compliance with the ethical standards of King Hussein Cancer Center for research involving human subjects and in accordance with the Declaration of Helsinki.
All patients diagnosed with early-stage, ER+, HER2- breast cancer who underwent ODx testing were included.
Demographic data were obtained from patients’ medical records. Histologic grade and biomarker data, including ER, PR, HER2, and Ki-67 were extracted when available; however, Ki-67 was not routinely reported in pathology reports during the earlier part of the study period at our institution, leading to incomplete availability. The study cohort was randomly divided into two subsets: a learning set and a validation set. Approximately two-thirds of the cases (n = 377; 71.4%) were assigned to the learning set, and the remaining one-third (n = 151; 28.6%) to the validation set.
Univariate analysis of the histopathologic and biomarker parameters in relation to the ODx score was carried out. The low-risk and high-risk scores were defined according to the TAILORx trial where a score of 0 - 25 is considered low-risk and 26 - 100 is considered high-risk. Descriptive statistics, t-test, Chi-square, receiver operating characteristics (ROC) curve, and univariate analysis of the histopathologic and biomarker parameters in relation to the ODx score were applied to the learning set to obtain an algorithm to predict RS category.
The sensitivity, specificity, positive predictive value, negative predictive value and accuracy were calculated to assess the performance of the algorithm in predicting low-risk or high-risk cases in the validation set.
| Results | ▴Top |
A total of 528 cases were included, with 377 patients assigned to the learning set and 151 to the validation set. Table 1 summarizes the pathological and clinical characteristics of the entire cohort. As shown in Table 2, there were no statistically significant differences between the learning and validation sets in terms of patient age, pathological features, or RS distribution, indicating that the two groups were comparable. In the learning cohort, 154 of 377 patients (40.8%) were under 50 years of age, and in the validation cohort, 60 of 151 patients (39.7%) were under 50. This is consistent with the reported proportion of breast cancer in women under 50 in Jordan as being 44% [32].
 Click to view | Table 1. Pathological Features for All Patients |
 Click to view | Table 2. Comparison of Various Variables Between the Learning Set and the Validation Set Showing No Significant Differences |
In the learning set, the RS risk category (low vs. high) was significantly associated with both histologic grade and the level of PR expression (Table 3). Grade 1 tumors were predominantly associated with low RS, whereas grade 3 tumors were more frequently associated with high RS. Similarly, higher levels of PR expression correlated with low RS, while lower PR expression levels were significantly associated with high RS.
 Click to view | Table 3. Relationship Between RS and Grade and RS and PR% in the Learning Set |
When we examined histologic grade in combination with PR percentage, we found a clear relationship with the RS (Table 3). All grade 1 had low RS regardless of the PR%, making them consistently low-risk.
For grade 2 and grade 3, the optimal PR% thresholds for predicting low-risk RS were determined using ROC curve analysis (Table 4). These cut-offs provided reasonable sensitivity and specificity, though not perfect.
 Click to view | Table 4. PR% Cut-Offs Combined With Grade as Determined by ROC Curves and Those Yielding a 100% Sensitivity |
To prioritize safety in clinical decision-making—specifically, to ensure that no high-risk cases are misclassified as low-risk—we adjusted the PR% thresholds to achieve 100% sensitivity. For grade 2 tumors, this threshold was determined to be ≥ 60% PR: among all grade 2 cases with PR ≥ 60%, none had a high-risk RS. However, at this threshold, the specificity was 69.1%, indicating that PR < 60% could not confidently rule out high risk, and these were therefore labeled undetermined.
For grade 3 tumors, a lower PR threshold of ≥ 40% achieved the same goal of 100% sensitivity. At this threshold, the specificity was 65%, and cases with PR < 40% were considered high risk, while those ≥ 40% were classified as undetermined due to limited predictive power. This approach emphasizes caution while maximizing the number of patients who can be confidently triaged. Notably, in grade 3 tumors, the model does not assign any patients directly to the low-risk category.
Based on these thresholds, approximately two-thirds of cases in the learning set could be confidently assigned to low- or high-risk categories, while 132 cases (35%) remained indeterminate (Fig. 1).
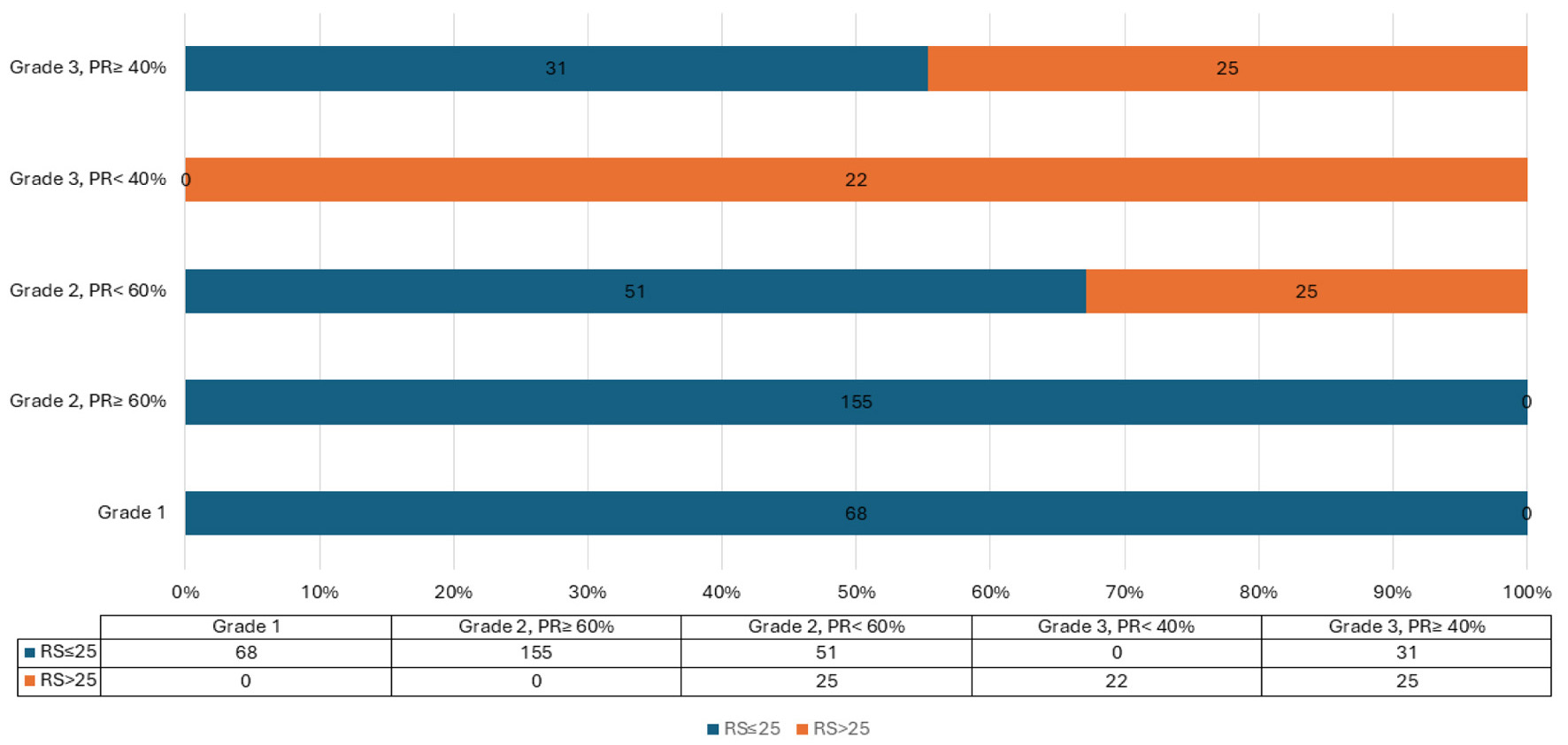 Click for large image | Figure 1. Proposed PR% and grade in relation to RS. PR: progesterone receptor; RS: recurrence score. |
An algorithm was developed (Fig. 2) using histologic grade and the proposed PR% cut-off values to predict RRS categories. This enables classification of approximately two-thirds of patients into a definitive risk group—low or high—based on grade and PR%, while identifying the remaining one-third as undetermined and likely to benefit from ODx testing. The algorithm was subsequently applied to the validation set. Figure 3 presents the results.
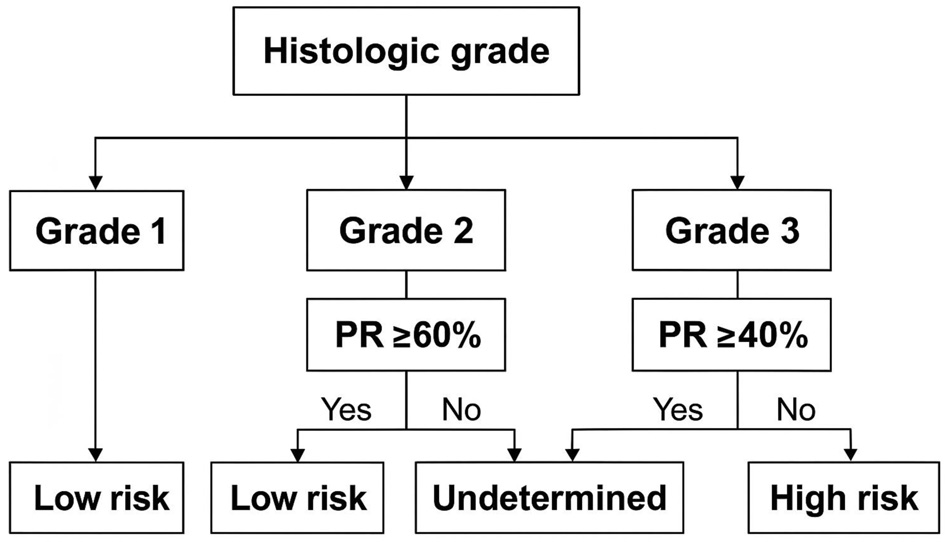 Click for large image | Figure 2. Proposed algorithm to predict Oncotype DX risk category using histologic grade and PR percentage. PR: progesterone receptor. |
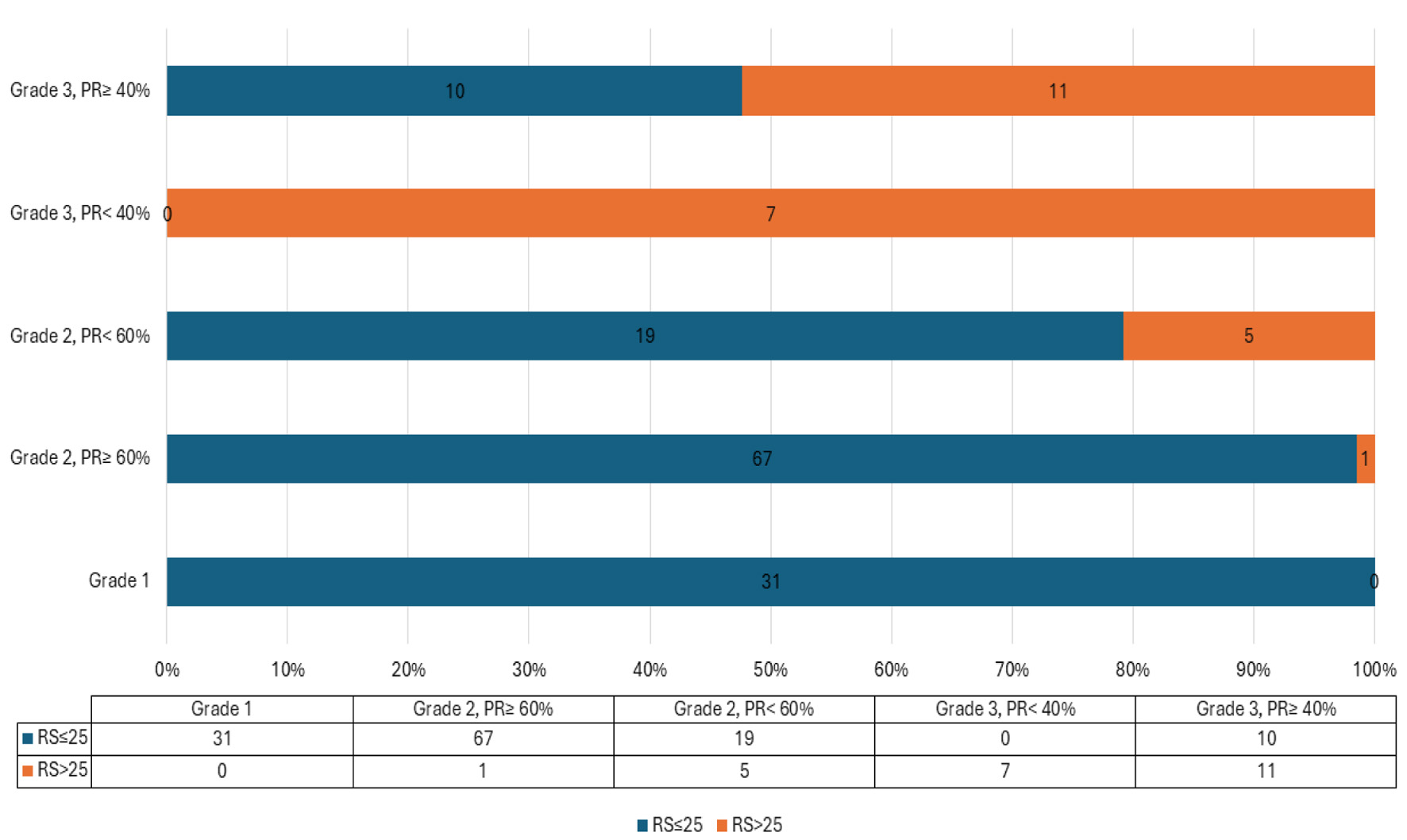 Click for large image | Figure 3. Results of applying the algorithm to the validation set. |
Based on the algorithm, cases were stratified into two groups: certain (those with confidently predicted RS category) and undetermined. In the validation cohort, the algorithm correctly classified all cases in the “certain” group into their appropriate RS categories, except for a single case in a grade 2 tumor with PR ≥ 60%. The undetermined group included 45 out of 151 cases (29.8%).
The algorithm demonstrated strong performance in predicting RS risk categories, with a sensitivity of 87.5%, specificity of 100%, positive predictive value (PPV) of 100%, negative predictive value (NPV) of 99%, and overall accuracy of 99%. The single misclassified case had RS of 26 which is just above the limit of the low score.
| Discussion | ▴Top |
Our study presents a feasible and robust model to predict the ODx RS risk category in ER+, HER2- early-stage breast cancer based on only two commonly reported parameters: histologic grade and percentage of PR positivity. Our data show that in this easy-to-use model, low-risk patients who are unlikely to derive benefit from chemotherapy might be identified safely, sparing the majority of patients from molecular testing.
Our findings match a growing number of studies that highlight the importance of histologic grade and PR status. A previous large-scale research by Orucevic et al has shown that these two factors are among the strongest predictors of RS, even more so than many complex clinicopathologic features in multivariable models [19]. Similarly, data from Canadian and Korean groups have reinforced the importance of grade and PR expression in estimating recurrence risk [18, 23]. More recently, Nozaki et al demonstrated that a model incorporating PR and Ki-67 effectively predicted recurrence risk as a surrogate for ODx [33]. However, in our cohort, Ki-67 was not routinely reported during the earlier part of the study period, leading to incomplete data availability. Moreover, Ki-67 is not universally assessed and is not recommended by current ASCO/CAP guidelines for treatment decisions, due to its poor reproducibility, lack of standardized thresholds, and considerable interobserver variability. These limitations reduce its reliability in daily practice and support the use of more consistently reported markers like histologic grade and PR%.
Several studies have also demonstrated a strong correlation between Ki-67 and tumor grade. Madani et al found that elevated Ki-67 levels were significantly associated with higher nuclear/histologic grades, HER2 positivity, and p53 expression [34]. Abubakr et al quantified this association, reporting a Spearman correlation coefficient of 0.68 (P < 0.001) and progressively increasing Ki-67 levels from grade 1 to grade 3 tumors [35]. Leksono et al further linked Ki-67 overexpression with higher tumor grade, larger tumor size, and poorer survival outcomes [36]. Together, these results suggest that histologic grade serves as a practical surrogate for tumor proliferative activity, capturing a significant portion of the prognostic insight offered by Ki-67.
While existing models often include these variables, they usually do so within multivariable equations or nomograms that need more data points, like Ki-67, tumor size, and LVI, along with external computation [25, 27, 29, 30]. This makes them less practical in real-world situations.
In comparison to these models, our algorithm eliminates such barriers. It uses only information that is universally available in breast cancer pathology reports and avoids any need for calculations or interpretation beyond what is immediately apparent. This approach proved highly effective.
In our cohort, grade 1 tumors were uniformly associated with low RS. Notably, this finding aligns with large-scale data indicating that high RS in grade 1 tumors is exceedingly rare. Multiple studies, including SEER-based analyses and validation of the AAMC model, have shown that fewer than 5%—and in some cases as few as 2.4%—of grade 1, ER+/HER2- tumors demonstrate a high RS [17, 20, 37-40]. These cases are so infrequent that their clinical significance is questionable, particularly as discordant cases (grade 1 with high RS) do not appear to derive substantial benefit from chemotherapy. Therefore, excluding a high-risk category for grade 1 tumors in our algorithm reflects both statistical rarity and limited therapeutic consequence [37].
For grade 2 and grade 3 tumors, PR thresholds of ≥ 60% and ≥ 40%, respectively, were sufficient to identify low-risk cases with high accuracy. Applying this algorithm, we were able to stratify approximately two-thirds of patients without requiring ODx testing, achieving 99% accuracy and 100% specificity in the validation cohort. Importantly, patients who could not be definitively classified were clearly flagged as “undetermined,” allowing clinicians to proceed with testing where appropriate.
At first glance, the PR thresholds used in our algorithm may seem unexpected—specifically, the use of a higher PR cut-off for grade 2 tumors (≥ 60%) compared to grade 3 tumors (≥ 40%). However, this distinction reflects differences in the intended purpose of the thresholds. For grade 2 tumors, the aim was to confidently identify patients with low-risk RSs; thus, a more stringent PR threshold (≥ 60%) was required to achieve 100% sensitivity and ensure that no high-risk cases were misclassified. Conversely, in grade 3 tumors, our model does not attempt to classify any patients as definitively low-risk. Instead, the threshold of 40% separates high-risk cases (PR < 40%) from an “undetermined” group (PR ≥ 40%), where the RS could still be low or high and ODx testing remains necessary. This cautious approach ensures that no potentially high-risk grade 3 tumors are misclassified while still reducing unnecessary testing in clearly high-risk cases. The absence of a low-risk arm for grade 3 tumors reflects the consistently elevated recurrence potential within this group, even when hormone receptor expression is preserved.
Our proposed approach demonstrates good diagnostic performance when compared with existing validated models, such as the Magee equations and Magee decision algorithm. The Magee equations which include ER/PR H-scores, Ki-67, mitotic score, and HER2 status as inputs have achieved sensitivity ranging from 89% to 94% and specificity ranging from 82% to 89% and with reports of up to 98% concordance between Magee-predicted and actual low-risk RS ODx in selected subgroups [10, 25]. The Magee decision algorithm, a combination of Magee equation scores and mitotic index, has been reported to achieve an overall accuracy of approximately 95% that safely identifies approximately 70% of women without the necessity of molecular confirmatory testing. However, this approach still requires quantitative ER/PR scoring and mitotic detail [27]. Similarly, the University of Tennessee nomogram, developed by Orucevic et al, uses multiple clinical and pathological variables—including grade, PR status (not percentage), tumor size, age, histologic type, and LVI—to predict RS category. While validated in over 84,000 patients and achieving a C-index of 0.81, its application typically depends on an online calculator or embedded software [19, 30]. Our model was able to confidently classify approximately two-thirds of patients using only histologic grade and PR percentage, without requiring any calculations, equations, or clinical inputs. This proportion is on par with the Magee algorithm and notably higher than other simpler models, such as the University of Chicago high-sensitivity rule-out approach, which safely classified only 43% of cases [22, 23]. Additionally, our model demonstrated a specificity of 100%, sensitivity of 87.5%, and an overall accuracy of 99% when tested on the validation cohort. While the sensitivity is slightly lower than that of certain Magee-based models, our approach offers a distinct advantage in specificity. This means no high-risk cases were mistakenly classified as low-risk, a critical factor in ensuring that patients receive appropriate treatment.
While many studies have recognized grade and PR status as key predictors of recurrence risk, our study is the first to propose a decision-based algorithm using specific PR percentage cut-offs rather than categorical status alone. For example, the models proposed by Gagnet et al and Thibodeau et al relied on binary classification of PR (positive vs. negative) without applying numeric thresholds limiting risk stratification [18, 22]. Our approach introduces clinically relevant cut-offs—PR ≥ 60% for grade 2 and ≥ 40% for grade 3—that enhance risk discrimination without adding complexity. Moreover, the structure of our model as a stepwise decision algorithm, rather than a formula or regression model, allows for immediate application in routine pathology practice.
While our findings highlight the usefulness of histologic grade and PR percentage in predicting RS, it is important to recognize that several studies have questioned whether clinicopathologic features can fully replace multigene assays. For example, research by Nitz et al [41], Stemmer et al [42], Sparano et al [3], and Bello et al [43] showed that traditional factors alone may not capture the full genomic complexity of breast cancer. This underscores the ongoing importance of multigene assays like ODx in guiding adjuvant therapy decisions. Still, in situations where genomic testing is not accessible or is cost-prohibitive, simplified models like ours can offer a practical way to triage patients—helping identify those who are very likely to have low-risk disease and may safely defer molecular testing. A robust, easy-to-use tool that accurately identifies low-risk patients using existing pathology data can support timely decision-making and reduce unnecessary delays. Even in high-resource settings, reducing unnecessary testing conserves healthcare resources and minimizes patient anxiety associated with waiting for genomic results.
This study is not without limitations. It was conducted retrospectively and in a single institution, which may affect generalizability. Our dataset reflects patients who elected to go for or could afford to undergo for ODx testing, which may introduce selection bias and limit generalizability, particularly for histologic subtypes that are less commonly tested. Additionally, PR assessment via immunohistochemistry can be subject to interobserver variability, particularly at percentage thresholds. Nonetheless, the consistency of our findings with prior studies, coupled with high performance in the validation cohort, supports the potential for broad applicability.
Additionally, approximately 40% of patients in both the learning and validation cohorts were under 50 years of age. This likely reflects real-world testing patterns, as clinicians are more likely to order genomic assays for younger patients due to concerns about undertreatment. We acknowledge that patient age influences treatment decisions for intermediate RS categories, particularly in women under 50 years old, as highlighted by the TAILORx trial [3]. However, our algorithm is designed to predict RS category, not to guide chemotherapy decisions directly. We maintained the conventional RS cut-off of < 25 for low risk to align with most clinical studies and assay standards. While chemotherapy may be considered for younger patients with RS 16 - 25 and high clinical risk, the RS itself remains a valid stratification metric. Importantly, the performance of our algorithm remained consistent across age groups, correctly predicting RS category in the majority of cases, thereby supporting its utility as a pre-test triage tool irrespective of age. Further stratification by age could be explored in future models aimed at tailoring treatment decisions rather than predicting RS category alone.
In conclusion, we propose a simple, practical algorithm that uses just two routinely reported pathology features—histologic grade and PR percentage—to guide treatment decisions in ER-positive, HER2-negative early breast cancer. By accurately identifying patients who are unlikely to benefit from chemotherapy, this model could help avoid unnecessary molecular testing in a large proportion of cases. While further validation in larger, multi-center studies will be important, our findings point to a valuable tool that can make clinical decision-making more efficient, especially in settings where resources are limited.
Acknowledgments
None to declare.
Financial Disclosure
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of Interest
The authors declare that there is no conflict of interest.
Informed Consent
Informed consent was waived due to the retrospective nature of the study and the use of de-identified data, in accordance with institutional policies.
Author Contributions
Maher A. Sughayer: conceptualization, supervision, methodology, and writing—original draft. Bayan Maraqa: data curation and writing—review and editing. Batool Qura’an: case selection, pathology data verification, and statistical analysis. Ahmad Alsughayer: surgical case data review and clinical validation. Hikmat Abdel-Razeq: clinical oversight, interpretation of findings, and manuscript revision. All authors approved the final manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article. Further details can be obtained from the corresponding author upon reasonable request.
| References | ▴Top |
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229-263.
doi pubmed - Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23(29):7350-7360.
doi pubmed - Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr., et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111-121.
doi pubmed - Andre F, Ismaila N, Allison KH, Barlow WE, Collyar DE, Damodaran S, Henry NL, et al. Biomarkers for adjuvant endocrine and chemotherapy in early-stage breast cancer: ASCO guideline update. J Clin Oncol. 2022;40(16):1816-1837.
doi pubmed - Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, Anderson B, et al. Breast Cancer, Version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(6):691-722.
doi pubmed - Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R. Histopathologic variables predict Oncotype DX recurrence score. Mod Pathol. 2008;21(10):1255-1261.
doi pubmed - Davey MG, Jalali A, Ryan EJ, McLaughlin RP, Sweeney KJ, Barry MK, Malone CM, et al. A novel surrogate nomogram capable of predicting OncotypeDX Recurrence Score(c). J Pers Med. 2022;12(7).
doi pubmed - Yoon KH, Lee SJ, Kim Y, Ahn JH, Kim JY, Park HS, Kim SI, et al. A simplified risk scoring system for predicting high-risk groups in gene expression tests for patients with estrogen receptor-positive, human epidermal growth factor receptor 2-negative, and node-positive breast cancer. Ann Surg Treat Res. 2023;105(6):360-368.
doi pubmed - Harowicz MR, Robinson TJ, Dinan MA, Saha A, Marks JR, Marcom PK, Mazurowski MA. Algorithms for prediction of the Oncotype DX recurrence score using clinicopathologic data: a review and comparison using an independent dataset. Breast Cancer Res Treat. 2017;162(1):1-10.
doi pubmed - Bhargava R, Clark BZ, Dabbs DJ. Breast cancers with magee equation score of less than 18, or 18-25 and mitosis score of 1, do not require oncotype DX testing: a value study. Am J Clin Pathol. 2019;151(3):316-323.
doi pubmed - Yamamoto S, Chishima T, Shibata Y, Harada F, Takeuchi H, Yamada A, Narui K, et al. Clinical impact of a novel model predictive of oncotype DX recurrence score in breast cancer. In Vivo. 2021;35(4):2439-2444.
doi pubmed - Lujan M, Moran D, Preciado BE, Lema C, Fernandez MJ, Egurrola JA, Pineda M, et al. Concordance between 21-gene recurrence score assay and clinicopathological predictive models in early-breast cancer patients cared for at a cancer center in Colombia. Gac Med Mex. 2023;159(1):3-9.
doi pubmed - Slembrouck L, Vanden Bempt I, Wildiers H, Smeets A, Van Rompuy AS, Van Ongeval C, Jongen L, et al. Concordance between results of inexpensive statistical models and multigene signatures in patients with ER+/HER2- early breast cancer. Mod Pathol. 2021;34(7):1297-1309.
doi pubmed - Patel R, Hovstadius M, Kier MW, Moshier EL, Zimmerman BS, Cascetta K, Jaffer S, et al. Correlation of the Ki67 Working Group prognostic risk categories with the Oncotype DX Recurrence Score in early breast cancer. Cancer. 2022;128(20):3602-3609.
doi pubmed - Dhungana A, Vannier A, Zhao F, Freeman JQ, Saha P, Sullivan M, Yao K, et al. Development and validation of a clinical breast cancer tool for accurate prediction of recurrence. NPJ Breast Cancer. 2024;10(1):46.
doi pubmed - Yoo SH, Kim TY, Kim M, Lee KH, Lee E, Lee HB, Moon HG, et al. Development of a nomogram to predict the recurrence score of 21-gene prediction assay in hormone receptor-positive early breast cancer. Clin Breast Cancer. 2020;20(2):98-107.e101.
doi pubmed - Lashen A, Toss MS, Fadhil W, Oni G, Madhusudan S, Rakha E. Evaluation oncotype DX((R)) 21-gene recurrence score and clinicopathological parameters: a single institutional experience. Histopathology. 2023;82(5):755-766.
doi pubmed - Gagnet S, Diorio C, Provencher L, Mbuya-Bienge C, Lapointe J, Morin C, Lemieux J, et al. Identifying clinicopathological factors associated with oncotype DX((R)) 21-gene recurrence score: a real-world retrospective cohort study of breast cancer patients in Quebec city, Canada. J Pers Med. 2021;11(9).
doi pubmed - Orucevic A, Bell JL, King M, McNabb AP, Heidel RE. Nomogram update based on TAILORx clinical trial results - Oncotype DX breast cancer recurrence score can be predicted using clinicopathologic data. Breast. 2019;46:116-125.
doi pubmed - Mohamed A, Kousar A, Wong J, Vohra N, Muzaffar M, Geradts J. Pathobiologic stratification of oncotype DX recurrence scores and comparative validation of 3 surrogate models. Arch Pathol Lab Med. 2022;146(10):1258-1267.
doi pubmed - Crager M, Wijayawardana SR, Gruver AM, Blacklock A, Russell C, Baehner FL, Sapunar F. Population-based estimate for the correlation of the Oncotype Dx Breast Recurrence Score(R) result and Ki-67 IHC MIB-1 pharmDx in HR+, HER2-, node-positive early breast cancer. Breast Cancer Res. 2022;24(1):74.
doi pubmed - Thibodeau S, Voutsadakis IA. Prediction of Oncotype Dx recurrence score using clinical parameters: A comparison of available tools and a simple predictor based on grade and progesterone receptor. Hematol Oncol Stem Cell Ther. 2019;12(2):89-96.
doi pubmed - Kim MC, Kwon SY, Choi JE, Kang SH, Bae YK. Prediction of oncotype DX recurrence score using clinicopathological variables in estrogen receptor-positive/human epidermal growth factor receptor 2-negative breast cancer. J Breast Cancer. 2023;26(2):105-116.
doi pubmed - Baltres A, Al Masry Z, Zemouri R, Valmary-Degano S, Arnould L, Zerhouni N, Devalland C. Prediction of Oncotype DX recurrence score using deep multi-layer perceptrons in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2020;27(5):1007-1016.
doi pubmed - Klein ME, Dabbs DJ, Shuai Y, Brufsky AM, Jankowitz R, Puhalla SL, Bhargava R. Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol. 2013;26(5):658-664.
doi pubmed - Singh K, He X, Kalife ET, Ehdaivand S, Wang Y, Sung CJ. Relationship of histologic grade and histologic subtype with oncotype Dx recurrence score; retrospective review of 863 breast cancer oncotype Dx results. Breast Cancer Res Treat. 2018;168(1):29-34.
doi pubmed - Bhargava R, Clark BZ, Carter GJ, Brufsky AM, Dabbs DJ. The healthcare value of the Magee Decision Algorithm: use of Magee Equations and mitosis score to safely forgo molecular testing in breast cancer. Mod Pathol. 2020;33(8):1563-1570.
doi pubmed - Turner BM, Finkelman BS, Hicks DG, Numbereye N, Moisini I, Dhakal A, Skinner K, et al. The Rochester Modified Magee Algorithm (RoMMa): an outcomes based strategy for clinical risk-assessment and risk-stratification in ER positive, HER2 negative breast cancer patients being considered for oncotype DX((R)) testing. Cancers (Basel). 2023;15(3).
doi pubmed - Bhargava R, Dabbs DJ. The story of the magee equations: the ultimate in applied immunohistochemistry. Appl Immunohistochem Mol Morphol. 2023;31(7):490-499.
doi pubmed - Orucevic A, Bell JL, McNabb AP, Heidel RE. Oncotype DX breast cancer recurrence score can be predicted with a novel nomogram using clinicopathologic data. Breast Cancer Res Treat. 2017;163(1):51-61.
doi pubmed - Ibrahim EM, Al-Quzi BA, Shaheen AY, Kulak MH, Refae AA, Al-Foheidi ME. Correlation between oncotype Dx recurrence score and PREDICT estimates in early breast cancer: a single institution experience. JCO Glob Oncol. 2024;10:e2400112.
doi pubmed - Abdel-Razeq H, Mansour A, Jaddan D. Breast cancer care in Jordan. JCO Glob Oncol. 2020;6:260-268.
doi pubmed - Nozaki Y, Horimoto Y, Semba R, Ueki Y, Ishizuka Y, Onagi H, Hayashi T, et al. Attempt to substitute the oncotype DX Breast Recurrence Score((R)) test by histopathological factors and MUC1 protein expression. Cancer Diagn Progn. 2024;4(4):464-469.
doi pubmed - Madani SH, Payandeh M, Sadeghi M, Motamed H, Sadeghi E. The correlation between Ki-67 with other prognostic factors in breast cancer: a study in Iranian patients. Indian J Med Paediatr Oncol. 2016;37(2):95-99.
doi pubmed - Abubakr A, Humayun S, Ali T, Khursheed S, Khan A, Khan S, Akbar A, et al. Correlation Between Ki-67 Expression and Tumor Grade in Breast Cancer: A Cross-Sectional Study. Cureus. 2024;16(12):e76239.
doi pubmed - Leksono RB, Thabry R, Prihantono P, Nahusuly F, Kasim F, Hamdani W, Pieter J, et al. Correlation between the expression of Ki67 and histopathological grade, tumor size, disease-free survival, and overall survival among breast cancer patients. Breast Dis. 2022;41(1):455-460.
doi pubmed - Sibia US, Mylander C, Martin T, Rosman M, Sanders TJ, Lee Y, Tafra L, et al. OncotypeDX Testing Does Not Benefit Patients with Estrogen and Progesterone Receptor Positive Grade 1 Breast Cancers: A TAILORx Validated Study. Hematol Oncol Stem Cell Ther. 2023;16(4):412-419.
doi pubmed - Allemand C, Valerio AC, Calvo MF, Izbizky G, Mc Lean I, Terrier F, Uriburu JL, et al. Impact of the 21-Gene Recurrence Score (Oncotype DX®) on adjuvant therapy decision-making: a collaborative multicenter cohort study from Argentina. Mastology. 2021;31:1-7.
- Gage MM, Mylander WC, Rosman M, Fujii T, Le Du F, Raghavendra A, Sinha AK, et al. Combined pathologic-genomic algorithm for early-stage breast cancer improves cost-effective use of the 21-gene recurrence score assay. Ann Oncol. 2018;29(5):1280-1285.
doi pubmed - Gudena VK, Causey L, Stuart D, Martini K. Performance of Oncotype Dx test in patients with T1bN0, grade 1, hormone receptor-positive, HER2-negative breast cancer. Journal of Clinical Oncology. 2019;37(suppl; abstr):e12043.
- Nitz U, Gluz O, Christgen M, Kates RE, Clemens M, Malter W, Nuding B, et al. Reducing chemotherapy use in clinically high-risk, genomically low-risk pN0 and pN1 early breast cancer patients: five-year data from the prospective, randomised phase 3 West German Study Group (WSG) PlanB trial. Breast Cancer Res Treat. 2017;165(3):573-583.
doi pubmed - Stemmer SM, Steiner M, Rizel S, Soussan-Gutman L, Ben-Baruch N, Bareket-Samish A, Geffen DB, et al. Clinical outcomes in patients with node-negative breast cancer treated based on the recurrence score results: evidence from a large prospectively designed registry. NPJ Breast Cancer. 2017;3:33.
doi pubmed - Bello DM, Russell C, McCullough D, Tierno M, Morrow M. Lymph node status in breast cancer does not predict tumor biology. Ann Surg Oncol. 2018;25(10):2884-2889.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.