| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 000, Number 000, July 2025, pages 000-000
Clinical Utility of Targeted Next-Generation Sequencing for Determining Human Epidermal Growth Factor Receptor 2 Status and Optimizing Targeted Therapy in Breast Cancer
Yoshimi Haraa, h, Kazuki Moroa, h, Hiroshi Ichikawaa, i, Junko Tsuchidaa, Haruka Uchidaa, Kana Narusea, Hiroko Otakea, Yasuo Obataa, Mika Sugaib, Yoshifumi Shimadaa, Jun Sakataa, Hajime Umezuc, Yu Koyamad, Shujiro Okudae, Kazuaki Takabea, f, g, Toshifumi Wakaia
aDivision of Digestive and General Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
bDivision of Medical Technology, Niigata University Graduate School of Health Sciences, Niigata, Japan
cDivision of Pathology, Niigata University Medical and Dental Hospital, Niigata, Japan
dDepartment of Nursing, Niigata University Graduate School of Health Sciences, Niigata, Japan
eDivision of Bioinformatics, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
fDepartment of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
gDepartment of Surgery, University at Buffalo Jacobs School of Medicine and Biomedical Sciences, The State University of New York, Buffalo, NY, USA
hThese author contributed equally to this article.
iCorresponding Author: Hiroshi Ichikawa, Division of Digestive and General Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata City, Niigata 951-8510, Japan
Manuscript submitted April 19, 2025, accepted June 21, 2025, published online July 8, 2025
Short title: Targeted NGS for Breast Cancer
doi: https://doi.org/10.14740/wjon2583
| Abstract | ▴Top |
Background: The development of targeted next-generation sequencing (NGS) technologies has contributed to precision medicine, as evidenced by the growing interest in evaluating human epidermal growth factor receptor 2 (HER2) expression status to treat unresectable/metastatic HER2-low breast cancer (BC). However, the concordance between erb-b2 receptor tyrosine kinase 2 (ERBB2) copy number alteration (CNA) and HER2 immunohistochemistry (IHC) has never been determined. The aim of this study was to evaluate the utility of targeted NGS for determining HER2 status and optimizing targeted therapies for BC.
Methods: ERBB2 CNAs were examined by targeted NGS in 41 formalin-fixed paraffin-embedded (FFPE) BC tissues. ERBB2 CNA was compared with HER2 status evaluated by IHC in tissue sections, which were identical to those subjected to targeted NGS, using the Ventana 4B5 antibody.
Results: The median fold changes (FCs) for ERBB2 CNAs in tumors with an IHC score of 3+, 2+, 1+, and 0 were 4.81, 1.49, 1.00, and 1.00, respectively. The difference in the FC for ERBB2 CNA according to HER2 status was statistically significant (P < 0.001). An FC greater than 1.0 for ERBB2 CNA was established as the cutoff value to differentiate between tumors with an IHC score of 3+, 2+, or 1+ and tumors with an IHC score of 0, on the basis of receiver operating characteristic curve analysis. The overall percent agreement, positive percent agreement, negative percent agreement, and Cohen’s kappa between ERBB2 CNA and HER2 status were 68.3%, 57.7%, 86.7%, and 0.39, respectively. The numbers of patients with mutations in ERBB2, estrogen receptor 1 (ESR1), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), serine/threonine kinase 1 (AKT1), and phosphatase and tensin homolog (PTEN) were 7, 3, 6, 1, and 5, respectively. Targeted NGS detected additional gene mutations and presented treatment options for seven of 22 patients (31.8%) with an FC of ERBB2 CNA = 1.00.
Conclusions: Targeted NGS has the potential in distinguishing HER2 IHC 3+, 2+, and 1+ tumors from IHC 0 in patients with BC; however, differentiating between HER2 IHC 1+ and 0 remains challenging. Additionally, targeted NGS may aid in the identification of actionable mutations, thereby contributing to the selection of optimal treatment strategies in BC management.
Keywords: Breast cancer; Copy number alteration; Human epidermal growth factor receptor 2; Next-generation sequencing; Precision medicine
| Introduction | ▴Top |
Breast cancer (BC) is the leading cause of cancer-related death among women worldwide, and the estimated number of deaths in the USA was 42,170 in 2025 [1]. Strategies for BC treatment based on biomarkers, including hormone receptors (HRs), human epidermal growth factor receptor 2 (HER2), and programmed cell death ligand 1 (PD-L1), have improved in terms of progression-free survival (PFS) and/or overall survival (OS) in the last decade [2-4]. However, treatment strategies consisting of conventional cytotoxic agents, molecularly targeted therapies, and immune checkpoint inhibitors have become more complex than ever before, especially for metastatic BC (mBC). Multiple biomarkers, including those tested in companion diagnostics, must be evaluated to select the optimal treatment from among multiple complex treatment strategies. More accurate and simplified biomarker assessments could improve the quality of BC care.
HER2-positive BC [5], defined as BC with erb-b2 receptor tyrosine kinase 2 (ERBB2) amplification and/or protein overexpression, is identified in 15-25% of all invasive BCs [5-7] and is diagnosed by immunohistochemistry (IHC) and/or fluorescence in situ hybridization (FISH). The evaluation of HER2 status is necessary for selecting anti-HER2 treatments, including trastuzumab and pertuzumab [8-11]. Recently, the DESTINY-Breast04 phase III trial demonstrated the remarkable activity of trastuzumab deruxtecan (T-DXd), an antibody-drug conjugate (ADC) that targets HER2, to substantially improve PFS and OS compared with traditional chemotherapy among patients with HER2-low expression, which is defined as IHC 2+/FISH negativity or an IHC score of 1+ [12]. The indication for T-DXd was expanded for not only HER2-positive BC but also triple-negative and HR-positive BCs with HER2-low expression, as determined by companion diagnostic tests using IHC with a proprietary anti-HER2 antibody named as the VENTANA ultraView pathway HER2 (4B5).
In addition to HER2 status, several important actionable mutations in the additional genes including ERBB2, estrogen receptor 1 (ESR1), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), serine/threonine kinase 1 (AKT1), and phosphatase and tensin homolog (PTEN) have become biomarkers for selecting candidates with mBC for novel targeted therapies. Recently, ERBB2-mutant BC was suggested to be sensitive to neratinib [13], a potent irreversible pan-ERBB2 inhibitor. Although neratinib is not currently indicated for BC treatment in Japan, some studies, including the phase II SUMMIT basket trial, are ongoing [13]. In 2023, elacestrant was approved for the treatment of patients with ESR1-mutant HR-positive mBC [14]. Capivasertib (an AKT inhibitor) was also approved for patients who present with mBC with PIK3CA, AKT1, or PTEN mutations [15]. Alpelisib (an PI3Kα-specific inhibitor) was approved for patients with PIK3CA-mutated, HR-positive, HER2-negative advanced BC who had received endocrine therapy previously [16]. Thus, drug therapies targeting these oncogenic pathways have become treatment options for early mBC [17]. In addition to HER2 and HR status, the evaluation of oncogenic mutations in these genes is essential for developing treatment strategies for early mBC.
The development of targeted next-generation sequencing (NGS) technologies could provide an opportunity to implement genomics in clinical practice and to reveal an overall picture of genetic abnormalities such as single nucleotide variations, copy number alterations (CNAs), chromosomal abnormalities, and fusion genes in a wide range of cancers [18]. ERBB2 is the key gene in BC, and thus confirming the agreement between ERBB2 CNA evaluated by targeted NGS and HER2 status determined by IHC is crucial in the use of targeted NGS to suggest the standard of care in mBC. Targeted NGS can also analyze multiple genes, which could reveal simultaneous treatment strategies for targeted therapy [18]. Due to complex treatment strategies dictated by an increasing number of biomarkers, targeted NGS has the potential to play a central role in selecting drug therapies for patients with mBC.
In this study, we investigated the concordance between ERBB2 CNA and HER2 status, including HER2-low expression, in BC via genomic profiles constructed using targeted NGS. In addition, we delineated the landscape of additional gene mutations in ERBB2, ESR1, PIK3CA, AKT1, and PTEN in relation to HER2 status to select optimal treatment strategies consisting of T-DXd and novel targeted therapies, including neratinib, elacestrant, capivasertib, and alpelisib, for mBC.
| Materials and Methods | ▴Top |
Patient tissue samples
We conducted a single-center retrospective study. This study was approved by the Institutional Review Board of Niigata University (G2020-0038, 2018-0137).
Among the 589 patients with BC who underwent breast surgery at Niigata University Medical and Dental Hospital between 2016 and 2020, we enrolled 41 patients who had not received any other treatment, including chemotherapy or hormone therapy. Informed consent was obtained from all participants. All samples were derived from surgical specimens from patients. Among the cohort, 11 patients were included in our previous targeted NGS study of BC [19]. One patient diagnosed with stage IV BC underwent surgical resection for local control. Tumor stage was evaluated according to the American Joint Committee on Cancer (eighth edition) [20] and the general rules for clinical and pathological recording of BC (18th edition) [21].
IHC
Archival tissue from formalin-fixed paraffin-embedded (FFPE) tumors obtained from surgical specimens was used for analysis. Four-micrometer-thick slides were prepared, and the tumor tissues were stained with hematoxylin and eosin. To ensure the presence of > 50% tumor content in each section, the tumor content on hematoxylin and eosin-stained slides for each study sample was evaluated by an independent pathologist. The levels of estrogen receptor (ER) (1D5, Nichirei, Tokyo, Japan), progesterone receptor (PgR) (A9621A, Nichirei, Tokyo, Japan), and HER2 (clone 4B5, Ventana Roche Diagnostics, Tokyo, Japan) were determined by IHC. IHC staining was assessed according to the American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) guidelines [22]. HER2 scores were given according to the following criteria: 3+ for moderate to strong complete or basolateral membrane staining in 10% or more invasive tumor cells; 2+ for weak to moderate complete or basolateral membrane staining in 10% or more invasive tumor cells; 1+ for weak membrane staining in 10% or more invasive tumor cells; and 0 for no staining overall or membrane staining in fewer than 10% invasive tumor cells. ERBB2 amplification was evaluated by FISH in patients with an IHC score of 2+. FISH was performed using a PathVysion HER2 DNA probe kit (Abbott Molecular, Inc., Illinois, USA), and gene amplification was evaluated according to the fluorescence signal ratio of ERBB2 to chromosome enumeration probe 17 (CEP17). FISH positivity with gene amplification was defined as an ERBB2-to-CEP17 ratio of at least 2.0. HER2 positivity was defined as an IHC score of 3+ or an IHC score of 2+ in addition to FISH positivity, while HER2-low was defined as IHC 2+/FISH negativity or an IHC score of 1+, and HER2 negativity was defined as an IHC score of 0 [23].
NGS
Genomic profiles of BC tissues were generated by targeted NGS (CANCERPLEX, KEW Inc., Cambridge, MA, USA) using 435 cancer-associated genes, as previously described [24]. First, genomic DNA was extracted from FFPE BC tissues. Next, DNA fragment libraries were enriched for the coding regions and selected introns of the 435 genes and were then subjected to the Illumina MiSeq and NextSeq platforms (Illumina, San Diego, CA, USA) for genomic sequencing with an average depth of × 500. We extracted information on ERBB2 CNAs and additional gene mutations defined as single nucleotide variants (SNVs) and short insertions and deletions (indels) in genes related to BC targeted therapies including ERBB2, ESR1, PIK3CA, AKT1, and PTEN. The pathogenicity of the extracted mutations was identified via the ClinVar [25] and OncoKB [26] databases. Targeted therapies were recommended for cases harboring additional gene mutations classified as OncoKB therapeutic evidence level 1 (FDA-approved drug) [27].
Statistical analysis
The Kruskal-Wallis test was performed to assess the significance of differences in continuous data among the multiple groups. The Games-Howell test was applied to the post hoc tests for multiple comparisons. The cutoff value of ERBB2 CNA was determined via receiver operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) indicated accuracy according to the following criteria: AUC > 0.9, high accuracy; 0.7 < AUC ≤ 0.9, moderate accuracy; and 0.5 < AUC ≤ 0.7, low accuracy [28]. The concordance between ERBB2 CNA and HER2 IHC status was evaluated using overall percent agreement (OPA), positive percent agreement (PPA), negative percent agreement (NPA), and Cohen’s kappa with 95% confidence intervals (CIs). The relative strength of agreement was assessed according to the following criteria: kappa > 0.8, almost perfect; 0.6 < kappa ≤ 0.8, substantial; 0.4 < kappa ≤ 0.6, moderate; 0.2 < kappa ≤ 0.4, fair; 0 ≤ kappa ≤ 0.2, slight; and kappa < 0, poor [29]. Statistical evaluations were performed using the IBM SPSS version 28.0 (SPSS Japan, Tokyo, Japan) and the R programming language and environment (version 4.3.1) [30]. All tests were two-sided, and a P value less than 0.05 indicated statistical significance.
| Results | ▴Top |
Histologic and molecular characteristics of the 41 patients
In all, 41 patients were enrolled in this study. The patient backgrounds at diagnosis are summarized in Table 1, and the histologic and molecular characteristics of the tumors in each patient are detailed in Supplementary Material 1 (wjon.elmerpub.com). Only 15 (37%) patients had T1 classification and only 22 (54%) patients did not present with lymph node metastasis (Table 1). We evaluated HER2 status by IHC and FISH in tumor tissue sections derived from FFPE blocks that were identical to those subjected to targeted NGS. HER2 positivity (IHC 3+ or IHC 2+/FISH positivity) was detected in the tumors of nine patients (22.0%), including seven (17.1%) with IHC 3+ and two (4.9%) with IHC 2+/FISH-positive tumors. HER2-low expression (IHC 2+/FISH negativity, IHC 1+) was detected in the tumors of 17 patients (41.5%), including two (4.9%) IHC 2+/FISH-negative and 15 (36.6%) IHC 1+ tumors. HER2 negativity (IHC 0) was detected in the tumors of the remaining 15 patients (36.6%) (Supplementary Material 1, wjon.elmerpub.com). All patients who were HR-positive were ER-positive. We compared the ERBB2 CNA according to HER2 IHC (Fig. 1). The median fold changes (FCs) (range) for ERBB2 CNAs in HER2 IHC 3+, 2+, 1+, and IHC 0 were 4.81 (range: 1.73 - 14.03), 1.49 (range: 1.00 - 1.81), 1.00 (range: 1.00 - 1.70), and 1.00 (range: 0.57 - 1.33), respectively (Fig. 1). The difference in the FC for ERBB2 CNA according to HER2 status was statistically significant (P < 0.001). However, there were no significant differences between the groups of HER2 3+ and HER2 0 (P = 0.081), HER2 IHC 2+ and HER2 IHC 0 (P = 0.300), and HER2 IHC 1+ and HER2 IHC 0 (P = 0.192) (Fig. 1).
 Click to view | Table 1. Patient Background |
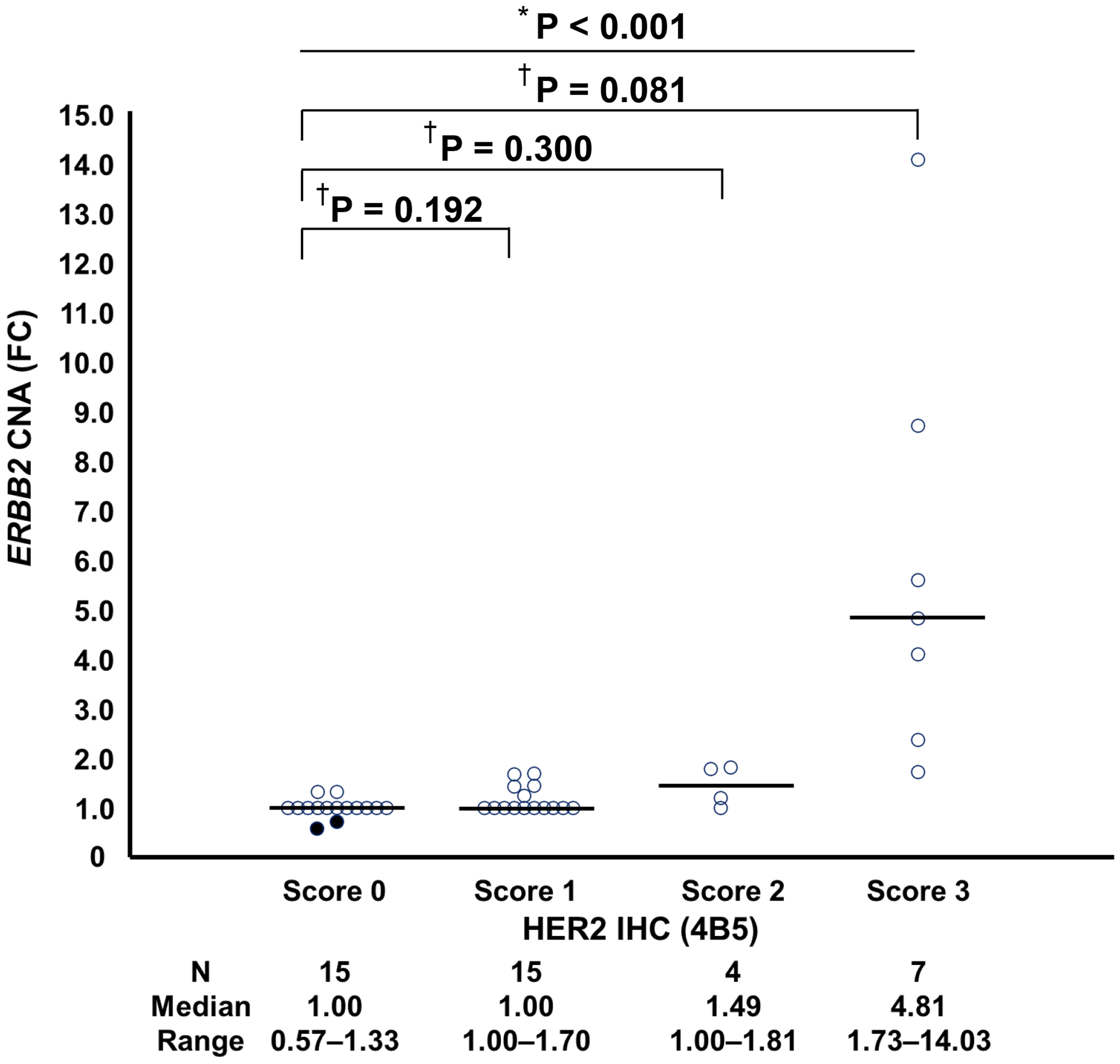 Click for large image | Figure 1. Distribution of ERBB2 CNAs according to HER2 IHC (4B5) expression status. Comparison of ERBB2 CNAs (FC) among patients with HER2 IHC (4B5) scores of 3+, 2+, 1+, and 0. The Kruskal-Wallis test was performed to assess the significance of differences in continuous data among the multiple groups (*). The Games-Howell test was applied to the post hoc tests for multiple comparisons (†). All tests were two-tailed, and P < 0.05 was considered significant. CNA: copy number alteration; ERBB2: erb-b2 receptor tyrosine kinase 2; FC: fold change; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry. |
Concordance between ERBB2 CNA evaluated by targeted NGS and HER2 status determined by IHC
To clarify the concordance between ERBB2 CNA evaluated by targeted NGS and HER2 IHC, we determined the cutoff value of ERBB2 CNA using receiver operating characteristic (ROC) curve analysis (Table 2, Supplementary Materials 2 and 3, wjon.elmerpub.com). ROC curve analysis revealed that FC of ERBB2 CNA over 1.00 (AUC = 0.78) was a fair discrimination point for distinguishing IHC scores of 3+, 2+, and 1+ from IHC 0. Fifteen of the 26 patients with an IHC score of 3+, 2+, or 1+ presented with an FC of ERBB2 CNA > 1.00, and the PPA was calculated to be 57.7% (Table 2). Among the 15 patients with an IHC score of 0, 13 patients presented an FC of ERBB2 CNA ≤ 1.00, and the NPA was calculated to be 86.7% (Table 2). The OPA was 68.3%, and fair agreement was determined with a Cohen’s kappa of 0.39 (95% CI: 0.14 - 0.64) (Table 2). We show representative examples of IHC based on ERBB2 CNA (Supplementary Material 4, wjon.elmerpub.com). No patients had an FC of ERBB2 CNA ≤ 1.00 for IHC 1+, an FC of CNA ≤ 1.00 for IHC 2+, an FC of CNA ≤ 1.00 for IHC 3+, or an FC of ERBB2 CNA = 1.00 for IHC 3+. As representative examples of concordance between the ERBB2 CNA, as evaluated by targeted NGS, and IHC, we identified the following patients: #1 (FC of ERBB2 CNA < 1.00, IHC 0), #4 (FC of ERBB2 CNA = 1.00, IHC 0), #26 (FC of ERBB2 CNA > 1.00, IHC 1+), #25 (FC of ERBB2 CNA > 1.00, IHC 2+), and #41 (FC of ERBB2 CNA > 1.00, IHC 2+). On the contrary, we show patient #14 (FC of ERBB2 CNA = 1.00, IHC 1+), patient #24 (FC of ERBB2 CNA = 1.00, IHC 2+), and patient #27 (FC of ERBB2 CNA > 1.00, IHC 0) as representative examples of discordance between ERBB2 CNA and HER2 IHC.
 Click to view | Table 2. Concordance Between ERBB2 CNA Evaluated by Targeted Next-Generation Sequencing and HER2 IHC (4B5) |
Landscape of additional gene mutations and proposed targeted therapies in relation to ERBB2 CNA
T-DXd and novel targeted therapies proposed for the enrolled patients according to ERBB2 CNA and additional gene mutations with ER status are shown in Figure 2. Lollipop plots depicting amino acid changes are presented in Figure 3, and a comprehensive summary of additional gene mutations along with corresponding treatment recommendations based on OncoKB therapeutic evidence levels is provided in Supplementary Material 5 (wjon.elmerpub.com). Additional gene mutations were detected in three of 17 patients (17.6%) with an FC of ERBB2 CNA > 1.00, in 15 of 22 patients (68.2%) with an FC of ERBB2 CNA = 1.00, and in neither of the two patients (0%) with an FC of ERBB2 CNA < 1.00.
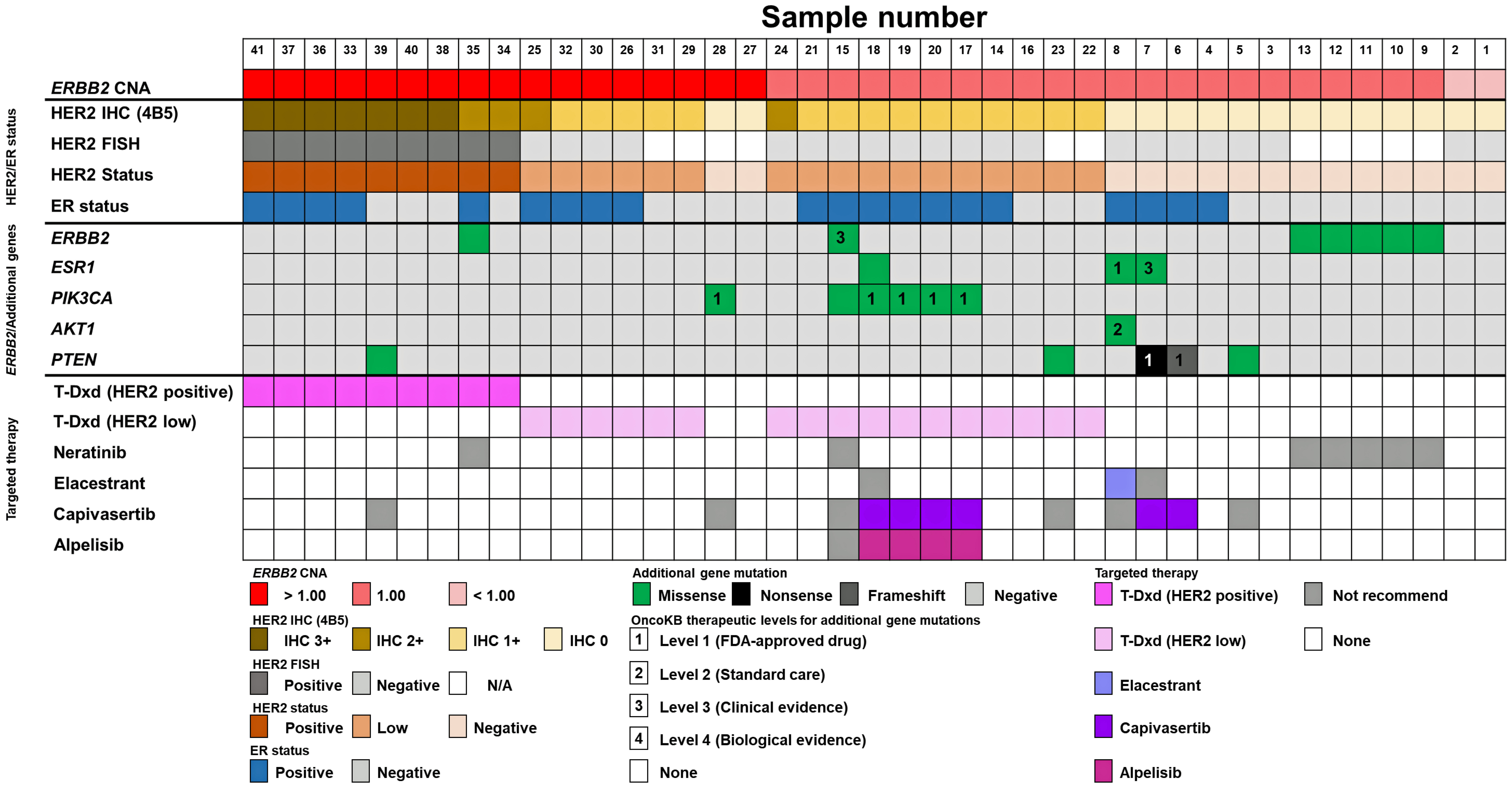 Click for large image | Figure 2. Landscape of additional gene mutations and proposed targeted therapies by targeted NGS, HER2, and ER status. ERBB2 CNA, HER2 status determined by HER2 IHC (4B5) and FISH, ER status, and additional gene mutations in ERBB2, ESR1, PIK3CA, AKT1, and PTEN along with the recommended targeted therapies based on OncoKB therapeutic evidence levels are depicted in color tiles. AKT1: serine/threonine kinase 1; CNA: copy number alteration; ER: estrogen receptor; ERBB2: erb-b2 receptor tyrosine kinase 2; FISH: fluorescence in situ hybridization; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry; NGS: next-generation sequencing; PTEN: phosphatase and tensin homolog. |
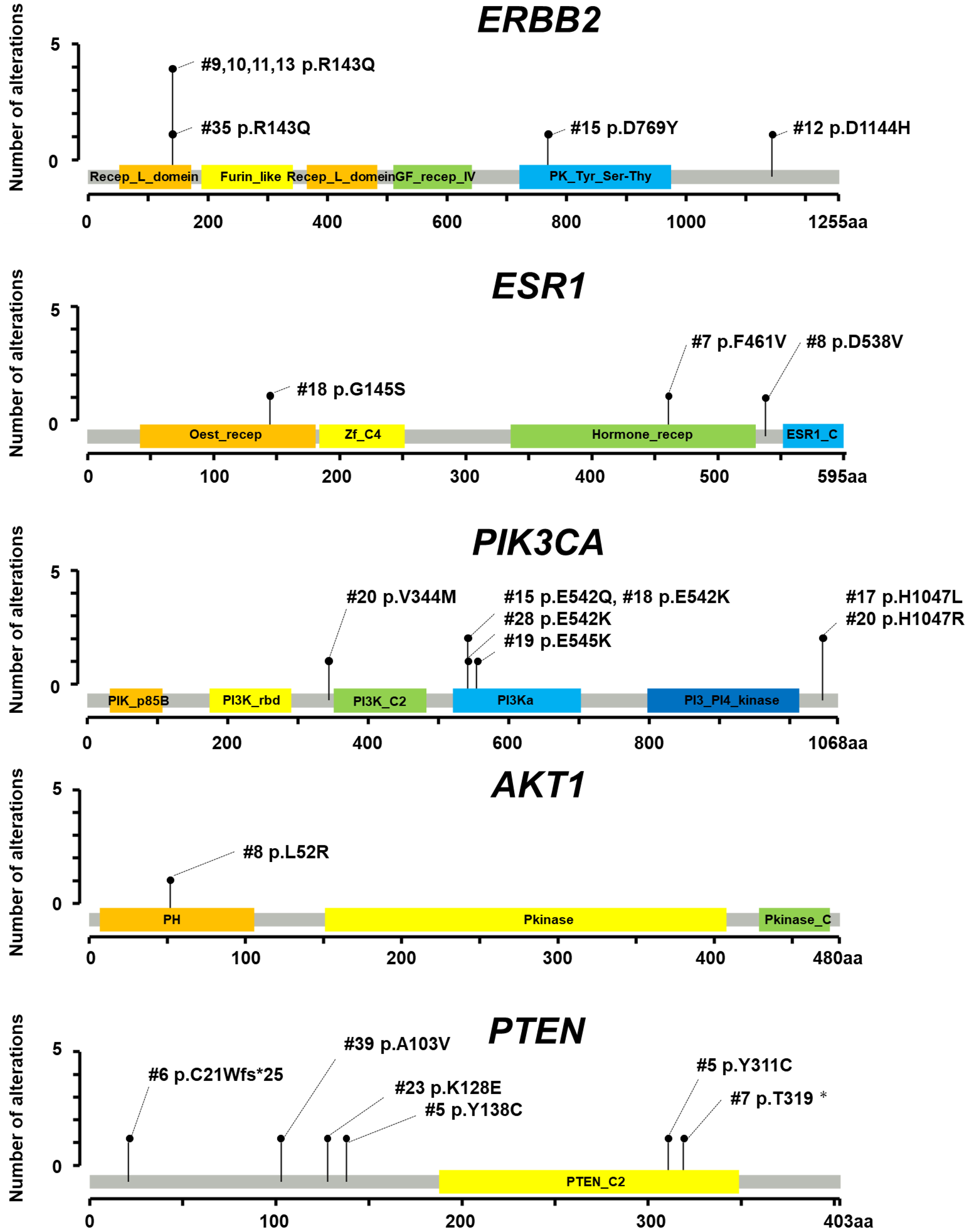 Click for large image | Figure 3. Mutations in ERBB2, ESR1, PIK3CA, AKT1, and PTEN detected in 41 patients. Lollipop plots, which show amino acid changes, are illustrated for ERBB2, ESR1, PIK3CA, AKT1, and PTEN. AKT1: serine/threonine kinase 1; ERBB2: erb-b2 receptor tyrosine kinase 2; ESR1: estrogen receptor 1; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PTEN: phosphatase and tensin homolog. |
Among the 17 patients with an FC of ERBB2 CNA > 1.00, one patient (#39) with a PTEN mutation and one patient (#35) with an ERBB2 mutation were not eligible for targeted therapy recommendations (Supplementary Material 5, wjon.elmerpub.com). Additionally, one patient (#28) with a PIK3CA E542K mutation was not considered for targeted therapy due to ER negativity. Overall, 15 patients (88.2%) with an FC of ERBB2 CNA > 1.00 were eligible for T-DXd treatment.
Among the 22 patients with an FC of ERBB2 CNA = 1.00, 15 patients (68.2%) harbored additional gene mutations and seven patients (31.8%) were recommended targeted therapies. Four patients (#17, #18, #19, and #20) with PIK3CA mutations were considered potential candidates for capivasertib and alpelisib. Although one patient (#18) also harbored an ESR1 mutation, elacestrant was not recommended; instead, capivasertib or alpelisib was proposed as a more suitable option. Two patients (#7 and #8) presented with ESR1 mutations along with either AKT1 or PTEN mutations. Of these, patient #7 was considered a candidate for capivasertib rather than elacestrant, whereas patient #8 was deemed a candidate for elacestrant over capivasertib according to OncoKB therapeutic evidence levels (Supplementary Material 5, wjon.elmerpub.com). One patient (#6) with a PTEN mutation was also considered a candidate for capivasertib. Conversely, no targeted therapies were recommended for one patient (#15) with ERBB2 and PIK3CA mutations and HER2-low, five patients (#9, #10, #11, #12, and #13) with ERBB2 mutations, and two patients (#5 and #23) with PTEN mutations (Supplementary Material 5, wjon.elmerpub.com). Additionally, two patients with an FC of ERBB2 CNA < 1.00 were not recommended any targeted therapy.
| Discussion | ▴Top |
Targeted NGS technologies, which can identify additional therapeutic targets in patients with recurrence or metastasis, have exploded on a worldwide scale. NGS libraries are prepared from only 100 ng of extracted tumor DNA and cover molecular target genes of BC, including ERBB2, ESR1, PIK3CA, AKT1, and PTEN [31]; however, HER2-low status is defined not by targeted NGS but by HER2 IHC. Although several reports have shown the genomic characterization of HER2-low BC via NGS [17, 32], the degree of concordance between ERBB2 CNA and HER2 status determined by IHC has never been examined. This is the first study to evaluate the degree of concordance between ERBB2 CNA determined by targeted NGS and HER2 status determined by IHC.
We demonstrated the distribution of ERBB2 CNAs determined by targeted NGS for different HER2 IHC scores (Fig. 1). Similar to some previous reports [32, 33], the median FC of ERBB2 CNAs tends to be higher in patients with higher HER2 IHC scores. In this study, an FC cutoff value of ERBB2 CNA > 1.00 demonstrated potential to distinguish HER2 IHC 3+, 2+, and 1+ tumors from IHC 0 in patients with BC; however, differentiation between HER2 IHC 1+ and HER2 0 remains challenging. As shown in Figure 1, two patients (#1 and #2) whose ERBB2 gene was deleted according to targeted NGS were determined to have an IHC score of 0. Considering that these two patients presented an FC of ERBB2 CNAs greater than 0.50 (Supplementary Material 1, wjon.elmerpub.com), the type of gene deletion might not be a homozygous deletion. Given that the frequency of ERBB2 single-copy deletions among HER2-negative BC patients was greater than that among HER2-low BC patients [31], our results also support this finding.
In this study, we showed that an FC of ERBB2 CNA ≤ 1.00 indicated an IHC score of 0, with a NPA of 86.7% (Table 2). This result indicated that patients with an FC of ERBB2 CNA > 1.00 might have an IHC score of 3+, 2+, or 1+, which would increase their chances of receiving T-DXd treatment. In contrast to the NPA, an FC of ERBB2 CNA > 1.00 indicated HER2 IHC scores of 3+, 2+, and 1+ with a PPA of 57.7%, which was lower than expected. Several HER2 IHC 1+ cases, potential candidates for T-DXd treatment, exhibited FC values of ≤ 1.00, potentially missing the opportunity for treatment. This raises the question of whether an FC cutoff value of ERBB2 CNA > 1.00 is truly optimal for distinguishing HER2 IHC 0 from higher expression levels (IHC 1+ to 3+). In contrast, we were able to effectively differentiate HER2 IHC 3+ or HER2 IHC 2+/FISH-positive tumors from others (Supplementary Material 2, wjon.elmerpub.com), and to distinguish the group of HER2 IHC 3+ or IHC 2+ from the group of HER2 IHC 1+ or IHC 0 using an FC cutoff of > 1.70, which demonstrated near-perfect concordance (Supplementary Material 3, wjon.elmerpub.com). Taken together, ERBB2 CNAs examined by targeted NGS may have limited utility in precisely identifying HER2-low status. Furthermore, confirmation of HER2 IHC expression may be necessary for tumors with ERBB2 CNA ≤ 1.00. However, IHC assessment is subject to variability due to staining conditions and inter-observer differences among pathologists, and thus cannot be considered an absolute standard. The optimal FC cutoff for ERBB2 CNA should be determined through further studies incorporating therapeutic outcomes, particularly the efficacy of T-DXd.
This inconsistent result may be explained by HER2 heterogeneity, which is defined as the presence of 5-50% cells with amplified ERBB2 or the presence of an area of the tumor that is HER2-negative [34], and increases the difficulty in evaluating HER2 expression status. Wu et al reported that HER2 IHC heterogeneity was more frequent in patients with HER2-low BC [35]. Moreover, since targeted NGS was performed on nucleic acids extracted from tumor tissues, it might be inappropriate to use targeted NGS to assess the heterogeneity of HER2 expression rather than HER2 IHC. The heterogeneity of HER2 expression might lead to discordance between ERBB2 can status and HER2 IHC scores. Taken together, although HER2 heterogeneity remains a concern, targeted NGS could indicate T-DXd treatment for patients with an FC of ERBB2 CNA > 1.00.
Although targeted NGS has raised concerns in the field of precision medicine, it has also presented opportunities to select optimal treatments for BC patients (Fig. 2). Interestingly, despite having ERBB2 CNA > 1.00, some patients received no recommendation for targeted therapy, even though T-DXd typically plays a central role in the treatment of mBC. In contrast, among patients with ERBB2 CNA = 1.00, seven of 22 (31.8%) were recommended targeted therapies. Notably, one patient (#8) harboring the ESR1 D538V mutation was identified as potentially responsive to elacestrant. ESR1 is one of the major drivers of endocrine resistance [36]. Elacestrant, the first oral selective estrogen receptor degrader (SERD), was approved for ESR1-mutant mBC in the USA. The phase III EMERALD study demonstrated that elacestrant was associated with longer median PFS than estrogen treatments in all patients (2.8 vs. 1.9 months for elacestrant and estrogen treatments, respectively), with a 45% relative risk reduction in disease progression or death in patients with the ESR1 mutation detected by Guardant 360 companion diagnostics (3.8 vs. 1.9 months, respectively) [14]. We utilized tumor biopsy rather than liquid biopsy for the detection of ESR1 mutation, in contrast to the approach used in the EMERALD study. Since prevalence of ESR1 mutation of liquid biopsy was higher than that of tumor biopsy [37], it is possible that the current study includes more cases with ESR1 mutations detected by liquid biopsy. Elacestrant therapy is a new effective option and has led to advancements in precision medicine for ER-positive and HER2-negative mBC, including HER2-low mBC.
Six patients (#6, #7, #17, #18, #19, and #20) harboring PIK3CA/AKT1/PTEN mutations were identified as potential candidates for capivasertib (Fig. 2). Overactivation of the PI3K-AKT-PTEN signaling pathway occurs in approximately half of HR-positive and HER2-negative BCs, including HER2-low BCs, and leads to worse outcomes [38, 39]. The CAPItello-291 study demonstrated that capivasertib plus fulvestrant was associated with a longer median PFS than fulvestrant alone (7.3 vs. 3.1 months) in mBC patients who were HR-positive, which is defined as ER positivity with or without PgR and HER2 negativity. The PIK3CA/AKT1/PTEN mutation group was defined as a molecular subgroup of tumors harboring at least one PIK3CA/AKT1/PTEN qualifying mutation [15]. In the CAPItello-291 study, at least one PIK3CA/AKT1/PTEN mutation was detected in 289/708 patients (40.8%). Mutations in PIK3CA only, AKT1 only, PTEN only, PIK3CA and AKT1, and PIK3CA and PTEN were detected in 202/708 (28.5%), 33/708 (4.7%), 37/708 (5.2%), 4/708 (0.6%), and 13/708 (1.8%) patients, respectively [15]. In this study, at least one PIK3CA/AKT1/PTEN mutation was detected in 8/15 patients (52.3%), while double gene mutations were not detected. Given that PIK3CA/AKT1/PTEN mutations were detected in 1,038/1,967 (52.8%) Japanese patients by FoundationOne [40], our results are expected. Four patients (#17, #18, #19, and #20) harboring PIK3CA mutations were identified as potential candidates for alpelisib therapy. The SOLAR-1 study demonstrated that alpelisib in combination with fulvestrant significantly prolonged median PFS than compared to fulvestrant alone (11.0 vs. 5.7 months) [16].
Thus, targeted NGS could be used to identify multiple targets and to develop strategies for cancer treatment. Moreover, since both targeted NGS and IHC are needed to select a treatment strategy for HR-positive mBC at present, further studies are needed to detect HER2-low BC by targeted NGS to avoid additional work needed to confirm each biomarker for drug therapy.
We acknowledge that the current study has limitations. First, we recognized cohort bias. Since we collected tissue samples without adversely affecting the diagnosis, we did not collect tissue samples from tumors < 10 mm in diameter or from cases of ductal carcinoma in situ. We also recognized the small cohort limited to Japanese patients. Second, although HER2 IHC heterogeneity was more common in HER2-low BC patients, we did not evaluate heterogeneity in this study or in the real world. Third, the targeted NGS utilized in this study lacks approval for clinical utilization worldwide. Nonetheless, we believe that our findings regarding the use of targeted NGS for BC are informative for cancer treatment.
In conclusion, targeted NGS has the potential in distinguishing HER2 IHC 3+, 2+, and 1+ tumors from IHC 0 in patients with BC; however, differentiating between HER2 IHC 1+ and 0 remains challenging. Additionally, targeted NGS may aid in the identification of actionable mutations, thereby contributing to the selection of optimal treatment strategies in BC management.
| Supplementary Material | ▴Top |
Suppl 1. Histologic and molecular characteristics of the tumors.
Suppl 2. Concordance between ERBB2 CNA evaluated by targeted next-generation sequencing and HER2 IHC (4B5).
Suppl 3. Concordance between ERBB2 CNA evaluated by targeted next-generation sequencing and HER2 IHC (4B5).
Suppl 4. Immunohistochemistry for HER2 (4B5). Representative images of HER2 IHC (4B5, × 400) in patients with ERBB2 CNAs with < 1, 1, and > 1 FC are shown. ERBB2, erb-b2 receptor tyrosine kinase 2; CNA, copy number alteration; FC, fold change; HER2, human epidermal growth factor receptor 2; IHC, immunohistochemistry.
Suppl 5. Additional gene mutations and treatment recommendation referred to OncoKB therapeutic evidence.
Acknowledgments
The authors greatly appreciate Akemi Kimoto for assisting with the immunohistochemical experiments, Miki Nitami for assisting with the management of research materials and funding, and Yukiko Kitamura for assisting with the literature searches and reviews.
Financial Disclosure
There was no specific funding source for this study.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper and that they approved the final version of the manuscript being submitted.
Informed Consent
Written informed consent for publication of individual patient data was obtained from the subjects for case presentation.
Author contributions
Y. Hara, K. Moro, H. Ichikawa, J. Tsuchida, and T. Wakai contributed to the conception and design of the study. K. Moro, H. Uchida, K. Naruse, H. Otake, Y. Obata, J. Tsuchida, and H. Ichikawa contributed to the collection and assembly of the data. M. Sugai and H. Umezu supervised the histopathological investigation. K. Moro, J. Tsuchida, H. Ichikawa, and S. Okuda performed the statistical and bioinformatic analyses. Y. Hara, K. Moro, J. Tsuchida, H. Ichikawa, Y. Shimada, J. Sakata, Y. Koyama, K. Takabe, and T. Wakai contributed to the drafting of the article. T. Wakai gave final approval for submission of the article.
Data Availability
The authors declare that the data supporting the findings of this study are available within the article. The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
AKT1: serine/threonine kinase 1; AUC: area under the ROC curve; BC: breast cancer; CDx: companion diagnostics; CNA: copy number alteration; EGFR: epidermal growth factor receptor; ER: estrogen receptor; ERBB2: erb-b2 receptor tyrosine kinase 2; ESR1: estrogen receptor 1; FC: fold change; FISH: fluorescence in situ hybridization; HER2: human epidermal growth factor receptor 2; IHC: immunohistochemistry; NGS: next-generation sequencing; NPA: negative percent agreement; OPA: overall percent agreement; PD-L1: programmed cell death ligand 1; PIK3CA: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PPA: positive percent agreement; PTEN: phosphatase and tensin homolog; ROC: receiver operating characteristic; SERD: selective estrogen receptor degrader
| References | ▴Top |
- Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75(1):10-45.
doi pubmed - Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Dieras V, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108-2121.
doi pubmed - Cortes J, Rugo HS, Cescon DW, Im SA, Yusof MM, Gallardo C, Lipatov O, et al. Pembrolizumab plus chemotherapy in advanced triple-negative breast cancer. N Engl J Med. 2022;387(3):217-226.
doi pubmed - Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, Denkert C, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382(9):810-821.
doi pubmed - Amodio R, Zarcone M, Cusimano R, Campisi I, Dolcemascolo C, Traina A, Agostara B, et al. Target therapy in HER2-overexpressing breast cancer patients. OMICS. 2011;15(6):363-367.
doi pubmed - Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14(4):320-368.
doi pubmed - Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177-182.
doi pubmed - Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200(3):290-297.
doi pubmed - Gianni L, Pienkowski T, Im YH, Roman L, Tseng LM, Liu MC, Lluch A, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25-32.
doi pubmed - Gianni L, Pienkowski T, Im YH, Tseng LM, Liu MC, Lluch A, Staroslawska E, et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): a multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016;17(6):791-800.
doi pubmed - Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, Ciruelos E, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724-734.
doi pubmed - Modi S. Trastuzumab deruxtecan in previously treated HER2-low advanced breast cancer: a plain language summary of the DESTINY-Breast04 study. Future Oncol. 2025;21(4):367-380.
doi pubmed - Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, Monsey J, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3(2):224-237.
doi pubmed - Bidard FC, Kaklamani VG, Neven P, Streich G, Montero AJ, Forget F, Mouret-Reynier MA, et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. J Clin Oncol. 2022;40(28):3246-3256.
doi pubmed - Turner NC, Oliveira M, Howell SJ, Dalenc F, Cortes J, Gomez Moreno HL, Hu X, et al. Capivasertib in hormone receptor-positive advanced breast cancer. N Engl J Med. 2023;388(22):2058-2070.
doi pubmed - Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929-1940.
doi pubmed - Schlam I, Chavez-MacGregor M. Best of the year: Advanced breast cancer in 2023. Breast. 2024;74:103677.
doi pubmed - Merlin JL, Husson M, Sahki N, Gilson P, Massard V, Harle A, Leroux A. Integrated molecular characterization of HER2-low breast cancer using Next Generation Sequencing (NGS). Biomedicines. 2023;11(12):3164.
doi pubmed - Nagahashi M, Ling Y, Hayashida T, Kitagawa Y, Futamura M, Yoshida K, Kuwayama T, et al. Actionable gene alterations in an Asian population with triple-negative breast cancer. JCO Precis Oncol. 2018;2.
doi pubmed - Giuliano AE, Connolly JL, Edge SB, Mittendorf EA, Rugo HS, Solin LJ, Weaver DL, et al. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(4):290-303.
doi pubmed - Tsuda H, General Rule Committee of the Japanese Breast Cancer S. Histological classification of breast tumors in the General Rules for Clinical and Pathological Recording of Breast Cancer (18th edition). Breast Cancer. 2020;27(3):309-321.
doi pubmed - Wolff AC, Somerfield MR, Dowsett M, Hammond MEH, Hayes DF, McShane LM, Saphner TJ, et al. Human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2023;147(9):993-1000.
doi pubmed - Ivanova M, Porta FM, D'Ercole M, Pescia C, Sajjadi E, Cursano G, De Camilli E, et al. Standardized pathology report for HER2 testing in compliance with 2023 ASCO/CAP updates and 2023 ESMO consensus statements on HER2-low breast cancer. Virchows Arch. 2024;484(1):3-14.
doi pubmed - Nagahashi M, Wakai T, Shimada Y, Ichikawa H, Kameyama H, Kobayashi T, Sakata J, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8(1):136.
doi pubmed - https://www.ncbi.nlm.nih.gov/clinvar/.
- https://www.oncokb.org/.
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1.
doi pubmed - Akobeng AK. Understanding diagnostic tests 3: Receiver operating characteristic curves. Acta Paediatr. 2007;96(5):644-647.
doi pubmed - Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159-174.
pubmed - http://www.r-project.org.
- Gilson P, Levy J, Rouyer M, Demange J, Husson M, Bonnet C, Salleron J, et al. Evaluation of 3 molecular-based assays for microsatellite instability detection in formalin-fixed tissues of patients with endometrial and colorectal cancers. Sci Rep. 2020;10(1):16386.
doi pubmed - Berrino E, Annaratone L, Bellomo SE, Ferrero G, Gagliardi A, Bragoni A, Grassini D, et al. Integrative genomic and transcriptomic analyses illuminate the ontology of HER2-low breast carcinomas. Genome Med. 2022;14(1):98.
doi pubmed - Tarantino P, Gupta H, Hughes ME, Files J, Strauss S, Kirkner G, Feeney AM, et al. Comprehensive genomic characterization of HER2-low and HER2-0 breast cancer. Nat Commun. 2023;14(1):7496.
doi pubmed - Hou Y, Nitta H, Li Z. HER2 intratumoral heterogeneity in breast cancer, an evolving concept. Cancers (Basel). 2023;15(10):2664.
doi pubmed - Wu Y, Zhong R, Ma F. HER2-low breast cancer: Novel detections and treatment advances. Crit Rev Oncol Hematol. 2023;181:103883.
doi pubmed - Herzog SK, Fuqua SAW. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br J Cancer. 2022;126(2):174-186.
doi pubmed - Sivakumar S, Jin DX, Tukachinsky H, Murugesan K, McGregor K, Danziger N, Pavlick D, et al. Tissue and liquid biopsy profiling reveal convergent tumor evolution and therapy evasion in breast cancer. Nat Commun. 2022;13(1):7495.
doi pubmed - Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016;2(12):1565-1573.
doi pubmed - Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
doi pubmed - Tada H, Miyashita M, Harada-Shoji N, Ebata A, Sato M, Motonari T, Yanagaki M, et al. Clinicopathogenomic analysis of PI3K/AKT/PTEN-altered luminal metastatic breast cancer in Japan. Breast Cancer. 2025;32(1):208-216.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.