| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 16, Number 1, February 2025, pages 16-29
Activity-Regulated Cytoskeleton-Associated Protein Gene Expression Is Associated With High Infiltration of Stromal Cells and Immune Cells, but With Less Cancer Cell Proliferation and Better Overall Survival in Estrogen Receptor-Positive/Human Epidermal Growth Factor Receptor 2-Negative Breast Cancers
Gabrielle Yeea, b, h , Rongrong Wua, c, h, Masanori Oshia, d, Itaru Endod, Takashi Ishikawac, Kazuaki Takabea, b, c, d, e, f, g, i
aBreast Surgery, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
bDepartment of Surgery, Jacobs School of Medicine and Biomedical Sciences, State University of New York, Buffalo, NY, USA
cDepartment of Breast Surgery and Oncology, Tokyo Medical University, Tokyo, Japan
dDepartment of Gastroenterological Surgery, Yokohama City University Graduate School of Medicine, Yokohama, Japan
eDepartment of Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
fDepartment of Breast Surgery, Fukushima Medical University School of Medicine, Fukushima, Japan
gDepartment of Immunology, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
hThese authors equally contributed to this manuscript.
iCorresponding Author: Kazuaki Takabe, Breast Surgery, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA
Manuscript submitted October 28, 2024, accepted January 3, 2025, published online January 13, 2025
Short title: ARC Gene Expression in ER+/HER2- BC
doi: https://doi.org/10.14740/wjon1936
| Abstract | ▴Top |
Background: Peritumoral lidocaine infiltration prior to excision is associated with better survival in breast cancer (BC), which led us to hypothesize that innervation to the tumor affects its biology and patient survival. Activity-regulated cytoskeleton-associated protein (ARC) gene expression is known to be regulated by neuronal activity. Therefore, we studied the clinical relevance of ARC gene expression as a surrogate of neuronal activity in BC.
Methods: Sweden Cancerome Analysis Network - Breast (SCAN-B (GSE96058), n = 3,273) cohort and The Cancer Genome Atlas (TCGA, n = 1,069) were analyzed.
Results: High ARC expression was significantly associated with smaller tumor size, without lymph node metastasis, and less stage IV disease in one cohort, but not validated by the other. Estrogen receptor-positive (ER+)/human epidermal growth factor receptor 2-negative (HER2-) and luminal A expressed significantly higher ARC compared to the other subtypes in both cohorts (P < 0.005). High ARC BC was significantly associated with lower Nottingham histological grade and lower Ki67 gene expression consistently in ER+/HER2- but not triple negative breast cancer (TNBC) in both cohorts (P < 0.001). Cell proliferation-related gene sets in the Hallmark collection (E2F targets, G2M checkpoint, and mitotic spindle) were significantly enriched to low ARC BC in ER+/HER2- but not TNBC in TCGA. The stromal cells (fibroblasts, vascular endothelial cells, and adipocytes) were all significantly infiltrated in high ARC ER+/HER2-, but not in TNBC, except for neurons. Homologous recombination deficiency, intratumor heterogeneity, fraction altered, silent or non-silent mutation rate were all significantly lower in high ARC ER+/HER2- but not TNBC. Although there was no difference in single nucleotide variant or indel neoantigens, tumor infiltrating lymphocytes, and cytolytic activity by ARC expression regardless of subtype, multiple immune cells were significantly infiltrated in high ARC ER+/HER2-, including CD8, CD4 memory cells, helper type II T cells, regulatory T cells, M2 macrophages, and B cells (all P < 0.03 in both cohorts), but not in TNBC. Disease-specific and overall survival were significantly improved in high ARC ER+/HER2- consistently in both cohorts (all P < 0.05), but this was not the case in TNBC.
Conclusion: ARC gene expression was associated with less cancer cell proliferation, high infiltration of stromal cells and immune cells, and better survival in the ER+/HER2- but not TNBC subtype.
Keywords: Activity-regulated cytoskeleton-associated protein; Breast cancer; Immunology; Molecular biology
| Introduction | ▴Top |
Breast cancer (BC) is the most common cancer worldwide and the leading cause of cancer death among women [1]. In the United States, BC accounts for 30% of all new female cancers each year. The incidence of BC has risen over the past four decades, a majority from earlier stage and hormone positive disease [2]. Several studies have shown that women diagnosed at an early stage have better survival and lower mortality rates [1, 3, 4]. While overall mortality has decreased, 30% of patients develop recurrent disease [5]. Therefore, the ability to study interventions that may affect future metastases is of importance.
Numerous studies have investigated factors associated with better biology [6, 7]. In the molecular era, the ability to use biomarkers for clinical treatment and prognostication for patients with BC is paramount [8]. Biomarkers such as estrogen receptor (ER), progesterone receptor, human epidermal growth factor receptor (HER) and Ki67 have been used extensively in clinics to determine hormone, anti-HER2, and chemotherapy treatment options [9, 10]. Indeed, we have recently shown the utility of Ki67 in identifying hepatocellular carcinomas with aggressive biology [11]. However, an increasing number of additional molecular biomarkers are being studied. Genes such as CACNG4, PKMYT1, EPYC, and CHRNA6 which play a role in cancer cell motility, the G2/M phase of cell cycles, extracellular matrix proteins, and neuronal nicotinic acetylcholine receptors, have been shown to be upregulated in BC patients compared to normal samples [12]. Our group has demonstrated that tumor infiltrating lymphocytes (TILs) in the bulk tumor are associated with cell proliferation and better survival in BC [13]. Additionally, high RAD51 gene expression, essential for homologous recombination of DNA repair, was found to be associated with aggressive cancer biology, cancer cell proliferation, and poor survival in BC [14]. Another marker of BC aggressiveness is the E2F score, which was predictive of the responsiveness of ER-positive/HER2-negative patients to neoadjuvant chemotherapy [15]. Many of these studies utilized The Cancer Genome Atlas (TCGA) cohort [16, 17] and Sweden Cancerome Analysis Network - Breast (SCAN-B, GSE96058) cohort [18, 19]. Both cohorts contain clinical parameters which are associated with full transcriptomes for every patient. Ultimately, these biomarkers have prognostic significance and may be targeted for future therapies.
Studies from the growing field of tumor neurobiology have demonstrated the role of nerves in cancer development and how nerves drive cancer initiation and progression. Many basic researchers are investigating the mechanisms by which cancer spreads with the use of nerves [20, 21]. Nerves can release neurotransmitters, hormones and growth factors, all of which can control cancer development and change the tumor microenvironment by promoting immune evasion and cancer progression [22, 23]. Innervation of tumors has been shown to play a critical part in head and neck, prostate, pancreatic, breast and gastric cancers [24]. Specifically, denervation of prostate tumors in mice models has been shown to inhibit cancer progression, and higher nerve density in the tumor microenvironment is associated with higher grade prostate cancer [25]. Similarly, BCs with a higher density of nerves have been found to be associated with poorer outcomes [26-28]. Denervating BC tumors in mice models has been shown to lead to volume regression [29]. While the exact molecular mechanism remains unclear, these insights are paving the way for interventions in immune-mediated diseases [30]. Recently, peritumoral lidocaine infiltration prior to surgical removal was reported to be associated with better survival in early-stage BC [31, 32], which demonstrates that nerves play a significant role in BC pathology, leaving opportunities to study the role of denervation on the tumor microenvironment.
In our modern day, the combination of clinical, pathological and molecular data allows us to gain information on prognostication and tumor microenvironment characterization [33]. The ability to create in silico models in computational biology has allowed us to identify numerous potential targets associated with BC. For example, when looking at the association of the angiopoietin pathway, a downstream angiogenesis cascade, high co-expression of the Ang2 gene and its receptor Tie2 were associated with decreased disease-free survival (DFS) and overall survival (OS) [34]. Not only can we identify clinically relevant gene expressions, but we can also determine RNA editing levels. We have shown that APOBEC3 enzymes, which contribute significantly to DNA mutagenesis in cancer, occur in BC tumors and are positively associated with increased immune activity and improved survival [35]. Additionally, the relationship between late recurrence and clinical factors, gene expression profiles and immune status has been studied. Late recurrence of BC has been shown to be associated with not only host defense immunity but also pro-cancerous immune cells and cytolytic activity of immune cells [36].
Activity-regulated cytoskeleton-associated protein (ARC) is a unique gene in neurons that is rapidly expressed in response to neural activity such as nerve stimulation [37]. ARC plays an essential role in synaptic transmission and long-term memory formation, interacting with effector proteins to modulate synaptic strength using molecular mechanisms [38]. ARC is a key regulator of synaptic plasticity, which is how synapses change in response to neural activity, impacting long-term potentiation and depression at the synapse level [39]. Although the link between ARC and neuronal modulation within the tumor microenvironment was never explored, ARC is known as a key molecular marker for recent neuronal activity and contributes to processes when a nerve is stimulated at the synapse level. ARC levels increase significantly at the stimulated synapse, allowing for structural changes that strengthen the connection between the neurons involved in that activity.
To this end, our overachieving hypothesis is that tumor innervation affects its biology and patient survival. In the current study, we utilized the ARC gene expression in the bulk tumor as a surrogate to estimate the neuronal activity in BC biology and investigated its clinical relevance in BC.
| Material and Methods | ▴Top |
BC bulk RNA sequencing cohorts
Two publicly available datasets were collected from online sources and analyzed in this study. As the testing cohort, we used the Pan Cancer Clinical Data Resource [40] and the cBio Cancer Genomic Portal [41] to obtain data on tumor gene expression and clinical data from TCGA cohort, which consists of over 11,000 patient samples, similarly to how we have previously reported [16, 17]. Among them, 1,069 patients with ER-positive/HER2-negative BC by pathological determination were analyzed. Nottingham histological grade of primary breast tumors of TCGA was assessed from pathology reports as shown previously [42]. The validation cohort used that has gene expression and correlating clinical data was the SCAN-B (n = 3,273), which was accessed using the cBio portal system that our group has previously described [18, 19].
All RNA sequence data utilized in this study were normalized and annotated to gene symbols from transcriptomics by the time of initial publication and followed by log2 conversion [18, 43]. Threshold values for defining “high” and “low” ARC expression were the median ARC gene expression in each cohort. All patient information included in both cohorts was de-identified; therefore, Institutional Review Board approval was not required for this study.
Single cell RNA sequencing cohorts of the primary BC
Three cohorts including single cell RNA seq data were downloaded from Gene Expression Omnibus (GEO) database [44] with the accession number of GSE255107 (n = 11), GSE167036 (n = 8), and GSE161529 (n = 52); two cohorts were downloaded from the Broad Institute Single Cell portal with the accession number of SCP1039 (n = 26) and SCP1106 (n = 5). A total of five cohorts were used for single cell sequencing analysis, using a cluster annotation method developed for single cells by the xCell research group [45-49]. These cohorts were downloaded from the GEO database via the R package GEOquary. GSE255107 is a cohort consisting of 11 primary tumors of patients with BC, including six ER-positive and five triple negative breast cancer (TNBC) [45]. GSE167036 consists of primary tumor and paired lymph node metastases in eight BC patients, five of which were luminal subtype and three HER2-positive [46]. GSE161529 contains data from 52 patients, four TNBC, four BRCA1 TNBC, six HER2-positive, 19 ER-positive, and six lymph node metastases of ER-positive tumors [47]. SCP1039 contains data from 26 primary tumors, including 11 ER-positive, five HER2-positive, and 10 TNBC [48]. Lastly, SCP1106 contains primary tumors from five patients with TNBC [49].
Gene Set Expression Analysis (GSEA)
We performed GSEA, a computational method to investigate the biological status of each set of genes [50], as performed in previous studies from our group [51, 52]. GSEA analyzes the statistical significance between two groups in the expression of pre-defined gene collection (gene set) representing specific biological pathways. Gene sets used in this study were from the Hallmark and Pathway Interaction Database (PID) collection of Molecular Signatures Database (MSigDB) [53]. The analysis was performed with GSEA software and hallmark gene set collection for pathway analysis. To compare results across gene sets, we used a normalized enrichment score (NES) adjusted for the correlation between the gene set and the expression dataset. Larger NES values indicate whether the pathway represented by each gene set is over- or under-enriched in each phenotype. We interpreted false discovery rate (FDR) q-values of 0.25 or less as statistically significant following the Broad Institute recommendations [50]. We used an FDR of 0.25 as previously recommended for GSEA.
Statistical analysis
The composition of infiltrating immune cells and stromal cells in the tumor microenvironment was estimated from gene expression data using xCell algorithm [54]. xCell is a web tool [55] that integrates the deconvolution approaches used in CIBERSORT [56], which is the most common method to dissect the tumor microenvironment using gene expression profiles, with the gene signature-based comparison method from GSEA [50]. This algorithm estimates cell type fractions by comparing 489 gene signatures corresponding to 64 cell types, including adaptive and innate immune cells, hematopoietic progenitor cells, epithelial cells, and extracellular matrix cells, with the input bulk tumor gene expression dataset.
Statistical analyses and data plotting were performed using R software and Microsoft Excel. A P-value threshold of 0.05 was used to detect statistical significance. To determine significance among different groups, we used one-way analysis of variance (ANOVA) or Fisher’s exact, as we described in legends. For survival analysis, the Kaplan-Meier method with log-rank test was used.
| Results | ▴Top |
ER-positive/HER2-negative and luminal A type, and T1 tumors have significantly high ARC expression
First, it was of interest whether ARC expression is related with clinical aggressiveness of BC, such as tumor size, lymph node metastasis, and subtypes. Thus, we analyzed the ARC expression by American Joint Committee on Cancer (AJCC) cancer staging and subtypes in ER-positive/HER2-negative and TNBC in the SCAN-B and TCGA cohorts. High ARC expression was significantly associated with smaller tumor size (Fig. 1a, P < 0.001) and with less lymph node metastasis in the SCAN-B cohort (Fig. 1a, P < 0.02); however, these results were not validated by the TCGA cohort. Although the SCAN-B cohort did not provide AJCC staging, high ARC expression was significantly associated with lower stage disease in the TCGA cohort (Fig. 1a, P < 0.02). ER-positive/HER2-negative and luminal A type cancers expressed significantly higher ARC compared to the other subtypes in both cohorts (Fig. 1b, P < 0.005).
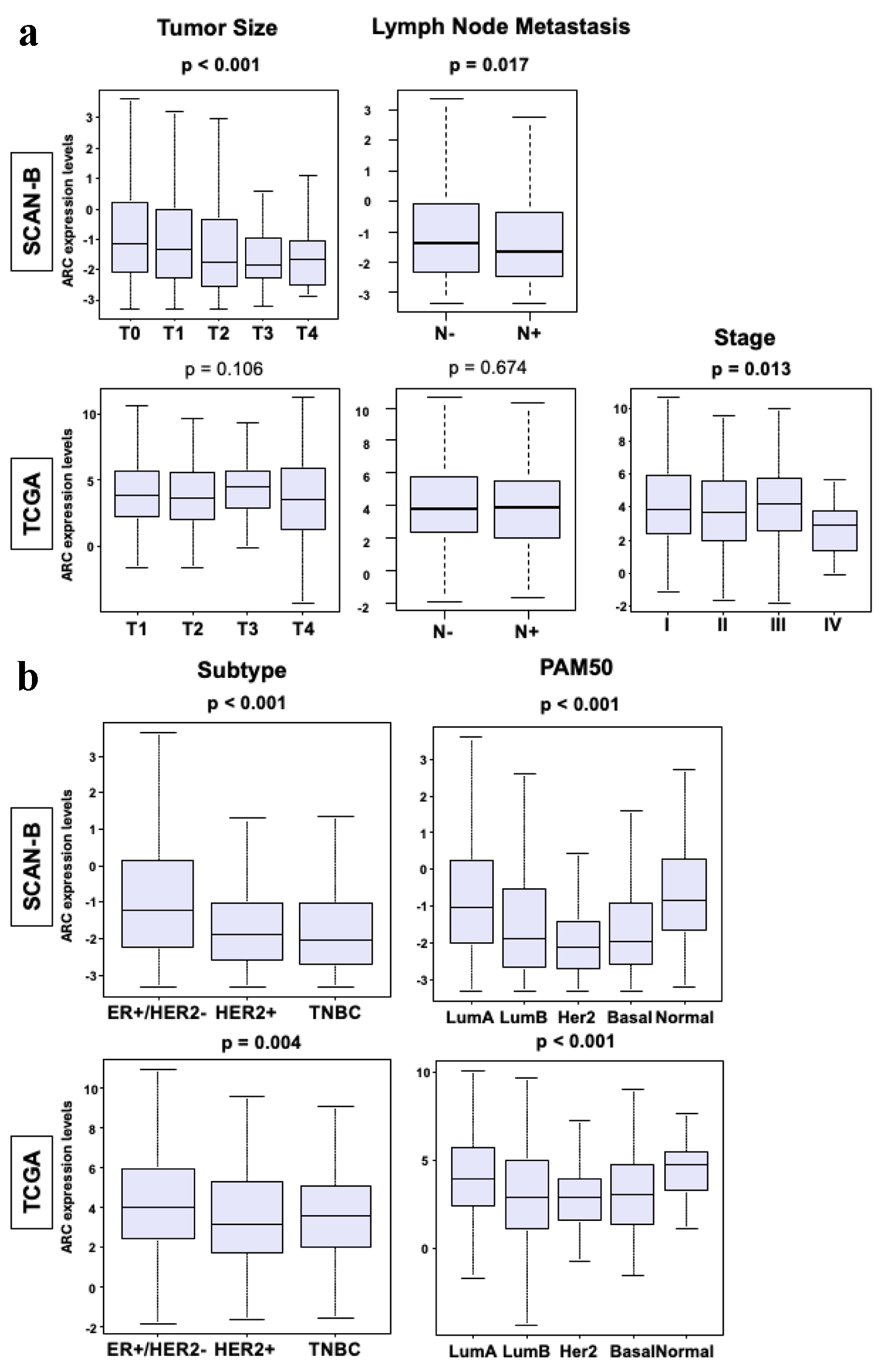 Click for large image | Figure 1. Clinical parameters of breast cancers in SCAN-B and TCGA cohorts. (a) Box plots of tumor size, lymph node metastasis and stage in the SCAN-B and TCGA cohorts. The Y-axis demonstrates ARC expression level. The X-axis represents tumor size (T1, T2, T3 and T4), patients with and without lymph node metastasis, and stage (I, II, III and IV) in each cohort. (b) Box plot of the subtypes of breast cancer in SCAN-B and TCGA cohorts. The X-axis represents ER-positive/HER2-negative, HER2-positive, TNBC, luminal A type, luminal B type, HER2-positive, basal type, or normal type. The Y-axis demonstrates ARC expression level. The Mann-Whitney U test and Fisher’s exact test were used to calculate P-values. ARC: activity-regulated cytoskeleton-associated protein; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; SCAN-B: Sweden Cancerome Analysis Network - Breast; TCGA: The Cancer Genome Atlas; TNBC: triple negative breast cancer. |
Cells with high ARC expression were lower grade, less aggressive, and less proliferative
Nottingham histological grade, which pathologically assesses cancer cell proliferation, was significantly lower in high ARC tumors in both TCGA and SCAN-B cohorts (Fig. 2a, P < 0.001). Gene expression of Ki67 (MKi67), one of the most common markers of cell proliferation in the clinical setting, was significantly lower in high ARC ER-positive/HER-2-negative tumors in both SCAN-B and TCGA (Fig. 2b, P < 0.001); however, this was not the case in the TNBC. Lastly, cell proliferation-related gene sets in the Hallmark collection (E2F targets, G2M checkpoint, and mitotic spindle) were all significantly enriched to low ARC BC in ER-positive/HER2-negative subtype, but none in TNBC subtype in TCGA cohort (Fig. 2c, all FDR < 0.25), but this result was not validated in SCAN-B cohort (data not shown, all FDR > 0.25).
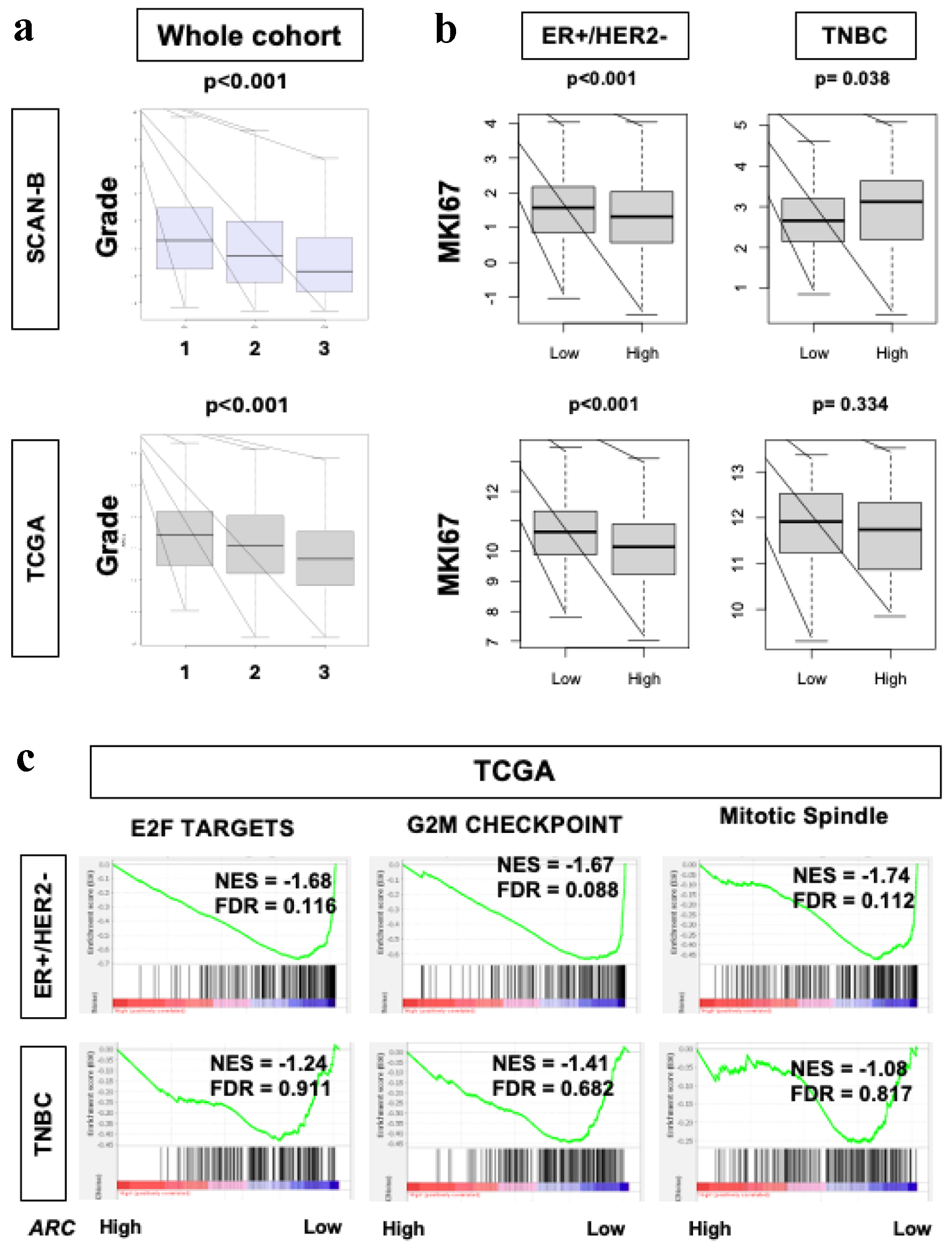 Click for large image | Figure 2. Cell proliferation rates in ER-positive/HER2-negative breast cancers in SCAN-B and TCGA. (a) Box plot demonstrating histological grade in the SCAN-B and TCGA cohorts. X-axis represents grade (1, 2 and 3). Fisher’s exact test was used to calculate P-values. (b) Box plots of MKi67 expression in SCAN-B and TCGA cohorts and within the ER-positive/HER2-negative and TNBC subgroups. Mann-Whitney U test was used to calculate P-values. (c) GSEA of Hallmark collection (E2F targets, G2M checkpoint, and MTOR1 signaling) and their relationship with ARC protein expression in SCAN-B and TCGA cohorts. Median cut-off was used to perform the analysis. NES and FDR were determined with the GSEA method. ARC: activity-regulated cytoskeleton-associated protein; ER: estrogen receptor; FDR: false discovery rate; GSEA: Gene Set Enrichment Analysis; HER2: human epidermal growth factor receptor 2; NES: normalized enrichment score; SCAN-B: Sweden Cancerome Analysis Network - Breast; TCGA: The Cancer Genome Atlas; TNBC: triple negative breast cancer. |
High ARC was associated with significantly more stromal cell infiltration and less mutation rates in ER-positive/HER2-negative but not TNBC
Previously we have shown that stromal cells such as adipocytes and lymphatic endothelial cells are highly infiltrated in a less proliferative tumor microenvironment [57-59]. The stromal cells (fibroblasts, endothelial cells, mvE and LyE cells, and adipocytes) were all found to be significantly infiltrated in high ARC ER-positive/HER2-negative BC tumor microenvironment in both cohorts (Fig. 3a, P < 0.02), but not TNBC (Fig. 3c) in either cohort. When looking at neurons specifically, there was significantly more infiltration of neurons in high ARC TNBC in both cohorts (Fig. 3c, both P < 0.01); however, this was not seen in the ER-positive/HER2-negative BC group in either cohort (Fig. 3a). Because mutation load is known to correlate with cancer aggression [60-62], we examined its association with ER-positive/HER2-negative and TNBC with low and high ARC. Using scores pre-calculated by Thorsson et al [63], we found that high ARC ER-positive/HER2-negative BC was significantly associated with less homologous recombination deficiency, intratumoral genomic heterogeneity, fraction altered mutation rate, silent and non-silent mutations compared to low ARC BC in TCGA (Fig. 3b, P < 0.04), but not TNBC (Fig. 3d). There was no difference in single nucleotide variant or indel neoantigens between tumors with low or high ARC expression in either ER-positive/HER2-negative or TNBC.
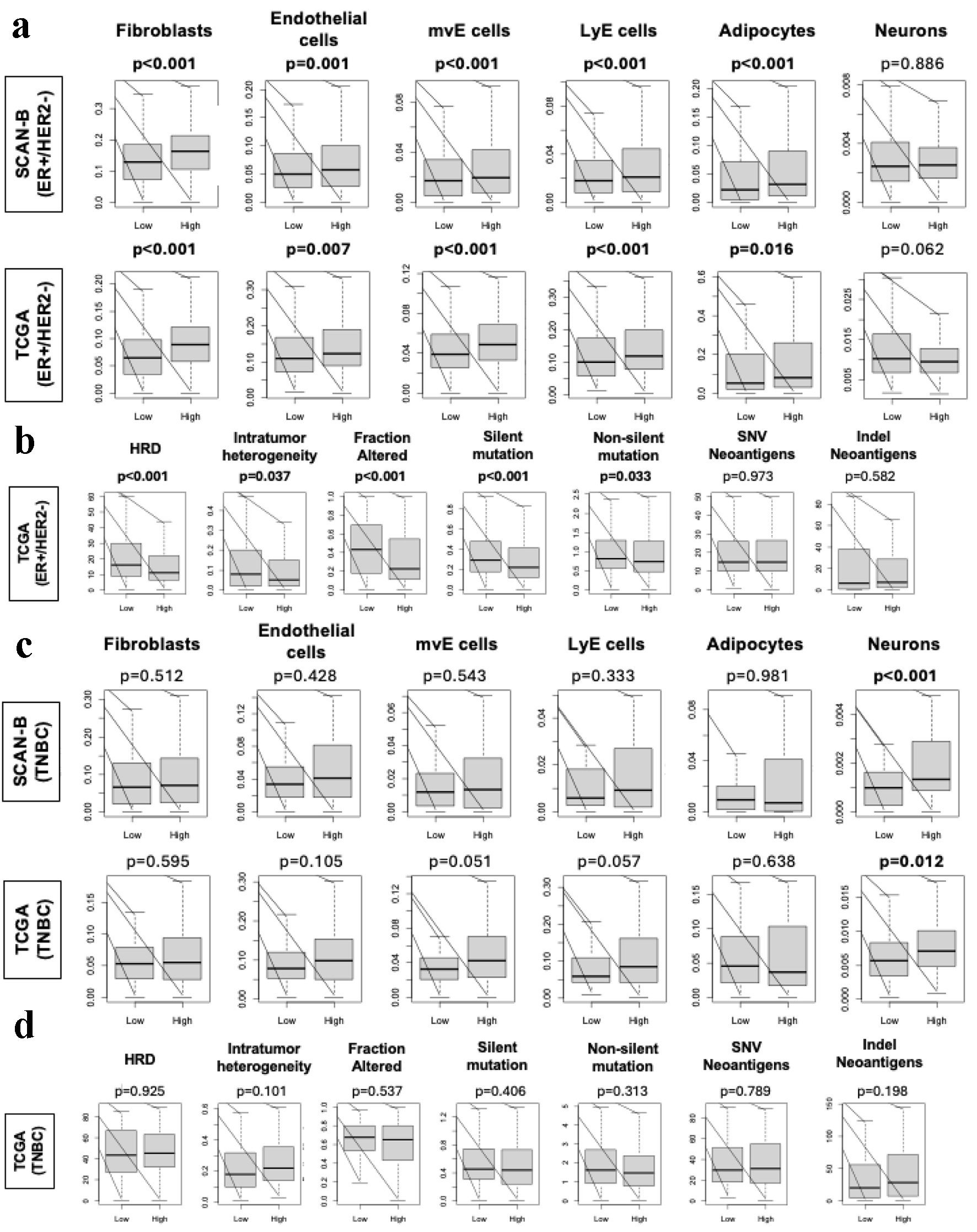 Click for large image | Figure 3. Stromal cell infiltration and mutation rates amongst ER-positive/HER2-negative and TNBC in SCAN-B and TCGA. (a) Box plots of stromal cells, including fibroblasts, endothelial cells, mvE cells, LyE cells, and adipocytes, and neurons in the ER-positive/HER2-negative patients in SCAN-B and TCGA cohorts. Box plot of stromal fraction based on Thorrson et al. (b) Box plots show HRD, intratumoral heterogeneity, fraction altered, silent and non-silent mutation rate, and SNV and indel neoantigens in the ER-positive/HER2-negative patients within the TCGA cohort based on scoring by Thorsson et al. P-values were determined using Mann-Whitney U test. (c) Box plots of stromal cells, including fibroblasts, endothelial cells, mvE cells, LyE cells, and adipocytes, and neurons in the TNBC patients in SCAN-B and TCGA cohorts. Box plot of stromal fraction based on Thorrson et al. (d) Box plots show HRD, intratumoral heterogeneity, fraction altered, silent and non-silent mutation rate, and SNV and indel neoantigens in the TNBC patients within the TCGA cohort based on scoring by Thorsson et al. P-values were determined using Mann-Whitney U test. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; HRD: homologous recombination deficiency; SCAN-B: Sweden Cancerome Analysis Network - Breast; SNV: single nucleotide variant; TCGA: The Cancer Genome Atlas; TNBC: triple negative breast cancer. |
High ARC expression in tumor microenvironment was associated with lower immune cell infiltration and less cytolytic activity in ER-positive/HER2-negative but not TNBC
We investigated the relationship of ER-positive/HER2-negative and TNBC tumors with ARC gene expression in the tumor microenvironment. We found that high ARC ER-positive/HER2-negative BCs were significantly associated with lower IFN-gamma response in TCGA (Fig. 4a, P < 0.01), but was not statistically significant in the TNBC group (Fig. 4a). TIL regional fraction score (Fig. 4a) nor cytolytic activity (Fig. 4b), which reflects overall anti-cancer immunity, was not statistically significant for either ER-positive/HER2-negative or TNBC groups. However, multiple immune cells were significantly infiltrated in high ARC ER-positive/HER2-negative BC, including CD8, CD4 memory cells, helper type II T cells, regulatory T cells, B cells, M2 macrophages, and dendritic cells (Fig. 4c, all P < 0.01 in both cohorts). In contrast, there was not as not much difference between immune cell infiltration between high and low ARC TNBC. The high ARC TNBC had significantly less infiltration of M1 macrophages (P < 0.03 for both cohorts), and significantly less CD8, CD4 memory, and B cells (P < 0.04 in SCAN-B) (Fig. 4d).
 Click for large image | Figure 4. Immune response in ER-positive/HER2-negative and TNBC tumor microenvironment. (a) Box plots of TILs regional fraction and INF-gamma response based on Thorrson et al in the ER-positive/HER2-negative and TNBC patients within the TCGA cohort. (b) Box plots of cytolytic activity in ER-positive/HER2-negative and TNBC patients within the SCAN-B and TCGA cohorts. (c) Box plots of immune cells, including CD8 cells, CD4 memory cells, Th1, Th2, Tregs, M1 and M2 macrophages, DCs, and B cells in the ER-positive/HER2-negative patients in the SCAN-B and TCGA cohorts. Mann-Whitney U test was used to determine P-values. (d) Box plots of immune cells, including CD8 cells, CD4 memory cells, Th1, Th2, Tregs, M1 and M2 macrophages, DCs, and B cells in the TNBC patients in the SCAN-B and TCGA cohorts. Mann-Whitney U test was used to determine P-values. DCs: dendritic cells; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; SCAN-B: Sweden Cancerome Analysis Network - Breast; TCGA: The Cancer Genome Atlas; Th1: T helper type 1; Th2: T helper type 2; TILs: tumor-infiltrating lymphocytes; TNBC: triple negative breast cancer; Tregs: regulatory T cells. |
Better OS was found in BCs with high ARC expression, particularly ER-positive/HER2-negative BC
Given the difference in tumor size, grade and AJCC stage between high and low ARC tumors, it was of interest to look at survival outcomes in the SCAN-B and TCGA cohorts. We found that OS was significantly better in high ARC BC in the ER-positive/HER2-negative subtype consistently in both cohorts (Fig. 5a, P < 0.01). However, no significant difference in OS or disease-specific survival (DSS) was found between low and high ARC gene expression in TNBC in either cohort (Fig. 5b). In fact, for TNBC, there was a trend of decreased OS and DSS for high ARC tumors in both SCAN-B and TNBC, but this was not statistically significant.
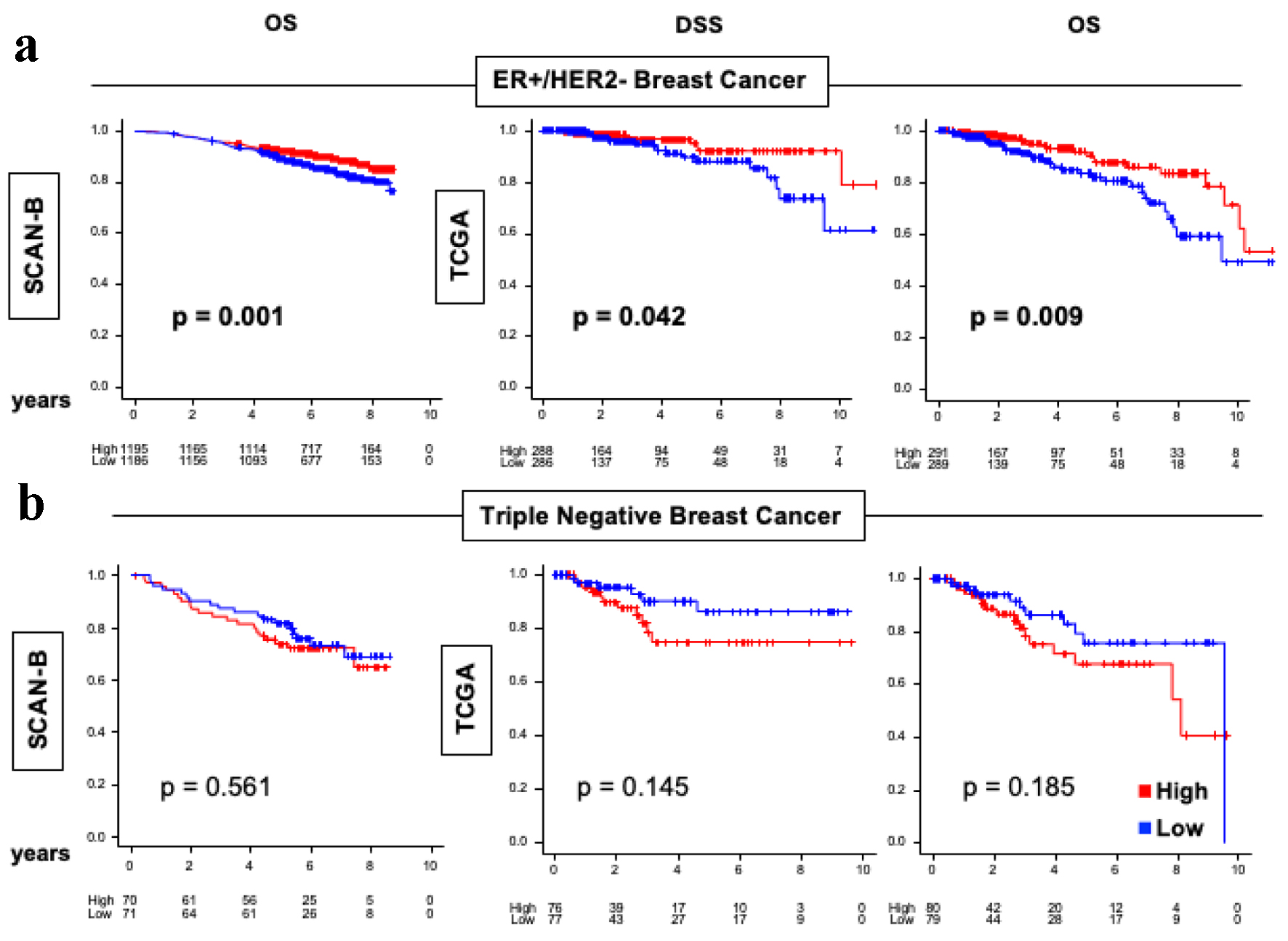 Click for large image | Figure 5. OS and DSS of ER-positive/HER2-negative and TNBC in the SCAN-B and TCGA cohorts. (a) Kaplan-Meier plots for OS in SCAN-B (n = 3,273) cohort and DSS and OS in TCGA (n = 1,069) cohorts within ER-positive/HER2-negative breast cancers. (b) Kaplan-Meier plots for OS in SCAN-B (n = 3,273) cohort and DSS and OS in TCGA (n = 1,069) cohorts within TNBC. Log-rank test was conducted to determine P-values. The red lines represent high ARC protein expression, and the blue lines represent low ARC protein expression. ARC: activity-regulated cytoskeleton-associated protein; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; OS: overall survival; DSS: disease-specific survival; SCAN-B: Sweden Cancerome Analysis Network - Breast; TCGA: The Cancer Genome Atlas; TNBC: triple negative breast cancer. |
Single cell analysis demonstrates inconsistent expression of ARC amongst cell types
Because ARC has been found to be expressed in neurons and involved with the synaptic transmission activity, it was of interest to see which cells within BC tumors express the ARC gene. On analysis of the single cell RNA sequencing results, the bulk of ARC expression was attributable to T cells in the ER-positive group and epithelial cells and neutrophils in the TNBC group in the GSE255107 cohort (Fig. 6a). In the GSE161529 cohort, ARC expression was attributable to epithelial and stem cells in both ER-positive and TNBC groups. Lastly in the SCP1039 cohort, ARC expression was attributable to normal epithelial cells in both ER-positive and TNBC groups. ARC expression was not preferentially expressed in any cell type in GSE167036 and SCP1106 (Fig. 6b).
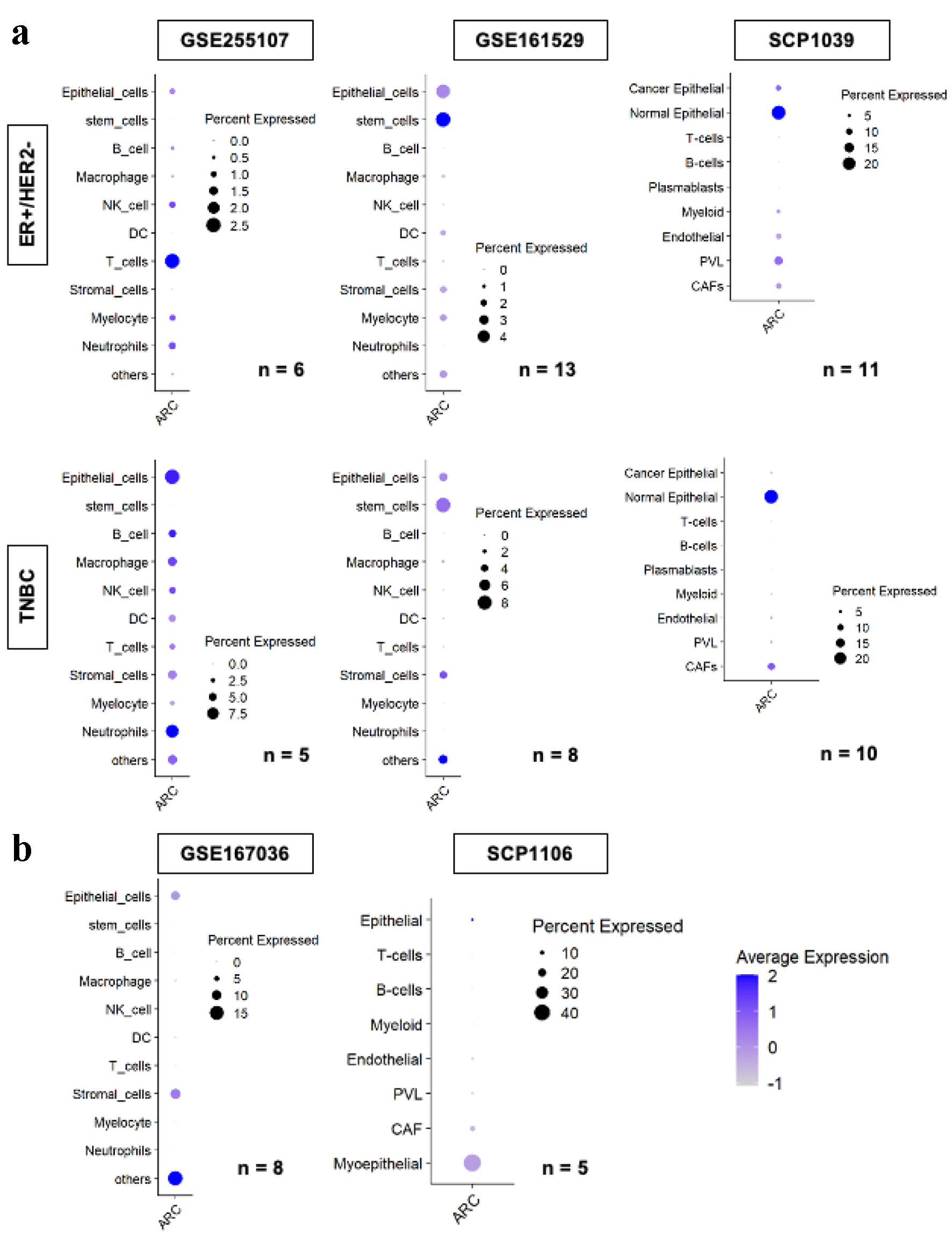 Click for large image | Figure 6. ARC gene expression by cell types in the tumor microenvironment amongst multiple single cell sequence cohorts. ARC expression of each cell type by subtype, ER+/HER2- in the upper row, and TNBC in the lower row, analyzed by single cell RNA sequencing in GSE255107, GSE161529, and SCP1039 cohorts (a) and GSE167036 and SCP1106 cohorts (b). Size of the circles indicates the percent of cells expressed ARC, and the darkness of the blue color indicates the average ARC expression in the cell type. “n” indicates the number of patients analyzed in the cohort. ARC: activity-regulated cytoskeleton-associated protein; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple negative breast cancer. |
| Discussion | ▴Top |
The aim of our study was to use ARC gene expression as a surrogate for neuronal activity and investigate the clinical relevance of its activity in the BC tumor microenvironment using SCAN-B and TCGA cohorts. When looking at the clinical parameters of tumors with high ARC, we found that high ARC expression was associated with ER-positive/HER2-negative and luminal A type BCs in both SCAN-B and TCGA cohorts. High expression of ARC was associated with smaller tumor size and lack of lymph node metastases in SCAN-B, but not in TCGA, most likely due to cohort sample size where SCAN-B has three times more power compared to TCGA. Furthermore, high ARC tumors were associated with lower grade and Ki67 gene expression in both SCAN-B and TCGA cohorts. We found that high ARC tumors were associated with lower Nottingham histological grade and lower Ki67 gene expression, both of which are well-established biomarkers of cancer cell proliferation, consistently in SCAN-B and TCGA cohorts. On the other hand, given that there was no consistent enrichment of Hallmark cell proliferation-related gene sets by ARC expression in the two cohorts, ARC expression may not be a strong biomarker to guide therapeutic decisions or prognostication in clinical practice, unlike the ones previously reported [8, 13, 52, 64]. There was no consistent enrichment of Hallmark cell proliferation-related gene sets by ARC expression in the two cohorts. We found that tumors expressing high ARC had significantly more stromal cells infiltrated and decreased mutation rates in ER-positive/HER2-negative cancers, but not TNBC. Because cancer aggressiveness is not only about the tumor itself but also its microenvironment, we analyzed the relationship between ARC expression and immune cells in the tumor microenvironment. ER-positive/HER2-negative cancers with high ARC had less immune cell infiltration and less cytolytic activity. OS was significantly higher with high ARC in ER-positive/HER2-negative BCs in both SCAN-B and TCGA, but not in TNBC. Surprisingly, ARC expression was inconsistently expressed amongst varying cell types in the tumor microenvironment when single cell analysis of ER-positive/HER2-negative and TNBC was analyzed.
There have been multiple studies reporting that innervation of a tumor plays an active role in its progression over the past decade. The elimination of specific nerves can halt the growth of a tumor depending on how that tumor is innervated in a particular tissue. Additionally, tumors themselves are likely to have a higher density of nerves compared to their original tissue. Because the innervation of tumors is not like that of normal tissue, it is of interest to understand how tumors become innervated in the first place and what relationship nerves may have with the molecular constitution of a tumor [65].
Sensory nerves that respond to pain may send damaging stimuli to the brain to help avoid future possible threats to an organism. Any type of harm to the body is usually associated with pain. More studies are showing that there is an overlap in pathways shared between nociceptors and the immune system which are playing a role in diseases and the body’s response to threats. For example, both immune cells and nociceptors release cell factors at the synaptic terminal in peripheral nerves of the spinal cord. The release of these factors plays a role in changing not only the immune system, but also the sensitivity of cells. It appears that this crosstalk between nociceptors and the immune system occurs at critical points in the body’s reaction to pain or inflammation [66].
Zhao et al found that denervation suppresses gastric tumorigenesis while studying all stages of gastric tumorigenesis in mouse models [67]. They showed that tumor stage correlated with neural density, and denervation by vagotomy decreased the risk of gastric cancer.
It is not clear what role nerves play in the BC tumor microenvironment. It is known that both sensory and sympathetic nerves exist in normal human breast tissue, contributing to the makeup of the skin and nipple, and blood vessels and ducts. As a result, it is possible that breast tumors also contain nerves, and more so than usual with the help of growth factors released by cancer cells. BC cells may secrete more than the usual amount of growth factors, thereby resulting in increased density of nerves within breast tumors [27].
Recently, a randomized clinical trial of peritumoral injection of lidocaine before BC surgery demonstrated significantly improved DFS and OS [31]. Given that lidocaine is one of the most commonly used local anesthetics, we cannot help but speculate that this result may implicate that numbing nerves to the tumor may suppress aggravation of the cancer cells during the resection of the tumor [32].
ARC is a gene coding for activity-regulated cytoskeleton-associated protein, which is a plasticity protein. ARC mRNA is derived from a retrotransposon [37] and is localized in activated synaptic sites, where the translated protein signaling via N-methyl-D-aspartate (NMDA) receptor [68, 69] plays a critical role in learning and memory-related molecular processes [70]. Dysfunction in the production of Arc protein can lead to various neurological disorders, such as Alzheimer’s disease [71] and amnesia [72]. ARC has been described as a critical regulator of dendritic cell migration in autoimmune diseases, and it was recently shown to enhance dendritic cell vaccination in experimental melanoma [73].
Our study showed that high ARC gene expression was associated with lower grade and less aggressive cancers with better survival. On the other hand, several studies have found that increased nerve density was associated with worse or invasive cancers. For example, Zhao et al found that neurogenesis was associated with aggressive features of invasive ductal carcinoma, tumor grade and patient survival, especially in ER-negative subtype, and not in ductal carcinoma in situ and fibroadenoma [74]. Pundavela et al similarly found that more nerve fibers were found in invasive ductal carcinoma compared to invasive lobular carcinoma (28% versus 12%), and even less so in ductal carcinoma in situ (8%) (P < 0.0003). Tumors that were lymph node positive had more nerve fibers than tumors that were lymph node negative (28% versus 15%, P < 0.0031), suggestive of metastatic potential [27]. In an analysis by Li et al, majority of the nerves in BC were sympathetic fibers. Patients with decreased survival were associated with tumors containing higher nerve density, regardless of lymph node metastasis [26]. Interestingly, Huang et al found that not just the presence of nerve fibers within BC tumors mattered, but also the thickness of the nerve. Higher stage tumors and those that were poorly differentiated were associated with thicker tumor nerve fibers. Interestingly, TNBC was also associated with thicker tumor nerve fibers [28]. While neurons clearly play a role in BC progression, it remains to be seen which subtype may benefit from neuronal intervention. Taken together, we cannot help but speculate that amount of nerve fibers is associated with aggressive BC, whereas ARC expression that reflects synaptic transmission is associated with less aggressive cancer.
Our study has some limitations. This is a retrospective study with possible selection bias, as the data were collected decades ago and do not include the prognostic impact of updated treatments. Additionally, sampling bias may exist due to the variations within bulk tumor samples because of the differences in proportion of the stromal area. Single-cell annotations are reflective of those used at the time of that cohort’s publication and are limited to single-cell annotation techniques at that time. Lastly, our data are derived in silico analysis and do not include experimental validation of the data. To this end, there is no proof of causality and exploration of a more detailed mechanistic link between ARC expression and cancer biology is expected to enhance understanding of role of tumor innervation in BC.
In conclusion, ARC gene expression as a surrogate of neuronal activity in BC was associated with high infiltration of stromal cells and immune cells but with less cancer cell proliferation and better OS, particularly in the ER-positive/HER2-negative subtype.
Acknowledgments
I, Gabrielle Yee, attest on behalf of all authors, that we had full access to the data of the study, conducted all data analyses independently from the funding entity, and take complete responsibility for the integrity and accuracy of the data reported in the manuscript. Part of this study was presented at the American Association for Cancer Research Meeting on April 8, 2024 in San Diego, California.
Financial Disclosure
This research was supported by National Institutes of Health, USA grant number R37CA248 -018, R01CA250412, R01CA251545, R01EB029596, as well as US Department of Defense BCRP, grant number W81XWH-19-1-0674 and W81XWH-19-1-0111, and National Institute of Health TO Roswell Park grant number P30CA016056 to K.T.
Conflict of Interest
The authors have no relevant financial or non-financial interests to disclose.
Informed Consent
Not applicable.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Rongrong Wu and Masanori Oshi. The first draft of the manuscript was written by Gabrielle Yee and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets analyzed in the current study were downloaded from cBioportal.
| References | ▴Top |
- Ong SK, Haruyama R, Yip CH, Ngan TT, Li J, Lai D, Zhang Y, et al. Feasibility of monitoring Global Breast Cancer Initiative Framework key performance indicators in 21 Asian National Cancer Centers Alliance member countries. EClinicalMedicine. 2024;67:102365.
doi pubmed - Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, Jemal A, et al. Breast cancer statistics, 2022. CA Cancer J Clin. 2022;72(6):524-541.
doi pubmed - Mittra I, Mishra GA, Dikshit RP, Gupta S, Kulkarni VY, Shaikh HKA, Shastri SS, et al. Effect of screening by clinical breast examination on breast cancer incidence and mortality after 20 years: prospective, cluster randomised controlled trial in Mumbai. BMJ. 2021;372:n256.
doi pubmed - Taylor C, McGale P, Probert J, Broggio J, Charman J, Darby SC, Kerr AJ, et al. Breast cancer mortality in 500 000 women with early invasive breast cancer diagnosed in England, 1993-2015: population based observational cohort study. BMJ. 2023;381:e074684.
doi pubmed - Courtney D, Davey MG, Moloney BM, Barry MK, Sweeney K, McLaughlin RP, Malone CM, et al. Breast cancer recurrence: factors impacting occurrence and survival. Ir J Med Sci. 2022;191(6):2501-2510.
doi pubmed - Hagerty BL, Takabe K. Biology of mesothelin and clinical implications: a review of existing literature. World J Oncol. 2023;14(5):340-349.
doi pubmed - Benesch MGK, Tang X, Brindley DN, Takabe K. Autotaxin and lysophosphatidate signaling: prime targets for mitigating therapy resistance in breast cancer. World J Oncol. 2024;15(1):1-13.
doi pubmed - Takabe K, Benesch MGK. Biomarker research in World Journal of Oncology. World J Oncol. 2023;14(1):1-3.
doi pubmed - Patani N, Martin LA, Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. Int J Cancer. 2013;133(1):1-13.
doi pubmed - Nishimukai A, Yagi T, Yanai A, Miyagawa Y, Enomoto Y, Murase K, Imamura M, et al. High Ki-67 expression and low progesterone receptor expression could independently lead to a worse prognosis for postmenopausal patients with estrogen receptor-positive and HER2-negative breast cancer. Clin Breast Cancer. 2015;15(3):204-211.
doi pubmed - Ramos-Santillan V, Oshi M, Nelson E, Endo I, Takabe K. High Ki67 Gene Expression Is Associated With Aggressive Phenotype in Hepatocellular Carcinoma. World J Oncol. 2024;15(2):257-267.
doi pubmed - Golestan A, Tahmasebi A, Maghsoodi N, Faraji SN, Irajie C, Ramezani A. Unveiling promising breast cancer biomarkers: an integrative approach combining bioinformatics analysis and experimental verification. BMC Cancer. 2024;24(1):155.
doi pubmed - Wu R, Oshi M, Asaoka M, Yan L, Benesch MGK, Khoury T, Nagahashi M, et al. Intratumoral tumor infiltrating lymphocytes (TILs) are associated with cell proliferation and better survival but not always with chemotherapy response in breast cancer. Ann Surg. 2023;278(4):587-597.
doi pubmed - Wu R, Patel A, Tokumaru Y, Asaoka M, Oshi M, Yan L, Ishikawa T, et al. High RAD51 gene expression is associated with aggressive biology and with poor survival in breast cancer. Breast Cancer Res Treat. 2022;193(1):49-63.
doi pubmed - Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, et al. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9(7):1-21.
doi pubmed - McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, Opyrchal M, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019;26(7):2191-2199.
doi pubmed - Katsuta E, Maawy AA, Yan L, Takabe K. High expression of bone morphogenetic protein (BMP) 6 and BMP7 are associated with higher immune cell infiltration and better survival in estrogen receptor-positive breast cancer. Oncol Rep. 2019;42(4):1413-1421.
doi pubmed - Saal LH, Vallon-Christersson J, Hakkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7(1):20.
doi pubmed - Oshi M, Gandhi S, Wu R, Asaoka M, Yan L, Yamada A, Yamamoto S, et al. Development of a novel BRCAness score that predicts response to PARP inhibitors. Biomark Res. 2022;10(1):80.
doi pubmed - Amit M, Na'ara S, Gil Z. Mechanisms of cancer dissemination along nerves. Nat Rev Cancer. 2016;16(6):399-408.
doi pubmed - Yaman I, Agac Cobanoglu D, Xie T, Ye Y, Amit M. Advances in understanding cancer-associated neurogenesis and its implications on the neuroimmune axis in cancer. Pharmacol Ther. 2022;239:108199.
doi pubmed - Khanmammadova N, Islam S, Sharma P, Amit M. Neuro-immune interactions and immuno-oncology. Trends Cancer. 2023;9(8):636-649.
doi pubmed - Hu J, Chen W, Shen L, Chen Z, Huang J. Crosstalk between the peripheral nervous system and breast cancer influences tumor progression. Biochim Biophys Acta Rev Cancer. 2022;1877(6):188828.
doi pubmed - Faulkner S, Jobling P, March B, Jiang CC, Hondermarck H. Tumor Neurobiology and the War of Nerves in Cancer. Cancer Discov. 2019;9(6):702-710.
doi pubmed - Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, Frenette PS. Autonomic nerve development contributes to prostate cancer progression. Science. 2013;341(6142):1236361.
doi pubmed - Li D, Hu LN, Zheng SM, La T, Wei LY, Zhang XJ, Zhang ZH, et al. High nerve density in breast cancer is associated with poor patient outcome. FASEB Bioadv. 2022;4(6):391-401.
doi pubmed - Pundavela J, Roselli S, Faulkner S, Attia J, Scott RJ, Thorne RF, Forbes JF, et al. Nerve fibers infiltrate the tumor microenvironment and are associated with nerve growth factor production and lymph node invasion in breast cancer. Mol Oncol. 2015;9(8):1626-1635.
doi pubmed - Huang D, Su S, Cui X, Shen X, Zeng Y, Wu W, Chen J, et al. Nerve fibers in breast cancer tissues indicate aggressive tumor progression. Medicine (Baltimore). 2014;93(27):e172.
doi pubmed - Kappos EA, Engels PE, Tremp M, Sieber PK, von Felten S, Madduri S, Meyer Zu Schwabedissen M, et al. Denervation leads to volume regression in breast cancer. J Plast Reconstr Aesthet Surg. 2018;71(6):833-839.
doi pubmed - Saraiva-Santos T, Zaninelli TH, Pinho-Ribeiro FA. Modulation of host immunity by sensory neurons. Trends Immunol. 2024;45(5):381-396.
doi pubmed - Badwe RA, Parmar V, Nair N, Joshi S, Hawaldar R, Pawar S, Kadayaprath G, et al. Effect of peritumoral infiltration of local anesthetic before surgery on survival in early breast cancer. J Clin Oncol. 2023;41(18):3318-3328.
doi pubmed - Chida K, Kanazawa H, Kinoshita H, Roy AM, Hakamada K, Takabe K. The role of lidocaine in cancer progression and patient survival. Pharmacol Ther. 2024;259:108654.
doi pubmed - Cai L, Xiao G, Gerber D, J DM, Xie Y. Lung Cancer Computational Biology and Resources. Cold Spring Harb Perspect Med. 2022;12(2):a038273.
doi pubmed - Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162(1):191-198.
doi pubmed - Asaoka M, Ishikawa T, Takabe K, Patnaik SK. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int J Mol Sci. 2019;20(22):1-16.
doi pubmed - Takeshita T, Yan L, Asaoka M, Rashid O, Takabe K. Late recurrence of breast cancer is associated with pro-cancerous immune microenvironment in the primary tumor. Sci Rep. 2019;9(1):16942.
doi pubmed - Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, et al. The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell. 2018;172(1-2):275-288.e218.
doi pubmed - Nikolaienko O, Patil S, Eriksen MS, Bramham CR. Arc protein: a flexible hub for synaptic plasticity and cognition. Semin Cell Dev Biol. 2018;77:33-42.
doi pubmed - Steward O, Worley PF. A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proc Natl Acad Sci U S A. 2001;98(13):7062-7068.
doi pubmed - Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e411.
doi pubmed - Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
doi pubmed - Takahashi H, Oshi M, Asaoka M, Yan L, Endo I, Takabe K. Molecular biological features of Nottingham histological grade 3 breast cancers. Ann Surg Oncol. 2020;27(11):4475-4485.
doi pubmed - Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70.
doi pubmed - https://www.ncbi.nlm.nih.gov/geo/.
- Tan J, Egelston CA, Guo W, Stark JM, Lee PP. STING signalling compensates for low tumour mutation burden to drive anti-tumour immunity. EBioMedicine. 2024;101:105035.
doi pubmed - Liu T, Liu C, Yan M, Zhang L, Zhang J, Xiao M, Li Z, et al. Single cell profiling of primary and paired metastatic lymph node tumors in breast cancer patients. Nat Commun. 2022;13(1):6823.
doi pubmed - Pal B, Chen Y, Vaillant F, Capaldo BD, Joyce R, Song X, Bryant VL, et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021;40(11):e107333.
doi pubmed - Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, Thennavan A, et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334-1347.
doi pubmed - Wu SZ, Roden DL, Wang C, Holliday H, Harvey K, Cazet AS, Murphy KJ, et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020;39(19):e104063.
doi pubmed - Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550.
doi pubmed - Sarkar J, Oshi M, Satyananda V, Chida K, Yan L, Maiti A, Hait N, et al. Spinster homologue 2 expression correlates with improved patient survival in hepatocellular carcinoma despite association with lymph-angiogenesis. World J Oncol. 2024;15(2):181-191.
doi pubmed - Takeshita T, Iwase H, Wu R, Ziazadeh D, Yan L, Takabe K. Development of a machine learning-based prognostic model for hormone receptor-positive breast cancer using nine-gene expression signature. World J Oncol. 2023;14(5):406-422.
doi pubmed - Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425.
doi pubmed - Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
doi pubmed - https://xcell.ucsf.edu/.
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-457.
doi pubmed - Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, et al. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21(16):5744.
doi pubmed - Mukhopadhyay S, Tokumaru Y, Oshi M, Endo I, Yoshida K, Takabe K. Low adipocyte hepatocellular carcinoma is associated with aggressive cancer biology and with worse survival. Am J Cancer Res. 2022;12(8):4028-4039.
pubmed - Wu R, Sarkar J, Tokumaru Y, Takabe Y, Oshi M, Asaoka M, Yan L, et al. Intratumoral lymphatic endothelial cell infiltration reflecting lymphangiogenesis is counterbalanced by immune responses and better cancer biology in the breast cancer tumor microenvironment. Am J Cancer Res. 2022;12(2):504-520.
pubmed - Chouliaras K, Oshi M, Asaoka M, Tokumaru Y, Khoury T, Endo I, Ishikawa T, et al. Increased intratumor heterogeneity, angiogenesis and epithelial to mesenchymal transition pathways in metaplastic breast cancer. Am J Cancer Res. 2021;11(9):4408-4420.
pubmed - Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, et al. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10(1):1852.
doi pubmed - Oshi M, Gandhi S, Huyser MR, Tokumaru Y, Yan L, Yamada A, Matsuyama R, et al. MELK expression in breast cancer is associated with infiltration of immune cell and pathological compete response (pCR) after neoadjuvant chemotherapy. Am J Cancer Res. 2021;11(9):4421-4437.
pubmed - Sayaman RW, Saad M, Thorsson V, Hu D, Hendrickx W, Roelands J, Porta-Pardo E, et al. Germline genetic contribution to the immune landscape of cancer. Immunity. 2021;54(2):367-386.e368.
doi pubmed - Oshi M, Wu R, Khoury T, Gandhi S, Yan L, Yamada A, Ishikawa T, et al. Infiltration of common myeloid progenitor (CMP) cells is associated with less aggressive tumor biology, lower risk of brain metastasis, better response to immunotherapy, and higher patient survival in breast cancer. Ann Surg. 2024;280(4):557-569.
doi pubmed - Reavis HD, Chen HI, Drapkin R. Tumor Innervation: Cancer Has Some Nerve. Trends Cancer. 2020;6(12):1059-1067.
doi pubmed - Pinho-Ribeiro FA, Verri WA, Jr., Chiu IM. Nociceptor sensory neuron-immune interactions in pain and inflammation. Trends Immunol. 2017;38(1):5-19.
doi pubmed - Zhao CM, Hayakawa Y, Kodama Y, Muthupalani S, Westphalen CB, Andersen GT, Flatberg A, et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med. 2014;6(250):250ra115.
doi pubmed - Wallace CS, Lyford GL, Worley PF, Steward O. Differential intracellular sorting of immediate early gene mRNAs depends on signals in the mRNA sequence. J Neurosci. 1998;18(1):26-35.
doi pubmed - Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30(1):227-240.
doi pubmed - McIntyre CK, Miyashita T, Setlow B, Marjon KD, Steward O, Guzowski JF, McGaugh JL. Memory-influencing intra-basolateral amygdala drug infusions modulate expression of Arc protein in the hippocampus. Proc Natl Acad Sci U S A. 2005;102(30):10718-10723.
doi pubmed - Yang L, Liu W, Shi L, Wu J, Zhang W, Chuang YA, Redding-Ochoa J, et al. NMDA receptor-arc signaling is required for memory updating and is disrupted in Alzheimer's disease. Biol Psychiatry. 2023;94(9):706-720.
doi pubmed - Gautam A, Wadhwa R, Thakur MK. Involvement of hippocampal Arc in amnesia and its recovery by alcoholic extract of Ashwagandha leaves. Neurobiol Learn Mem. 2013;106:177-184.
doi pubmed - Zhang XW, Huck K, Jahne K, Cichon F, Sonner JK, Ufer F, Bauer S, et al. Activity-regulated cytoskeleton-associated protein/activity-regulated gene 3.1 (Arc/Arg3.1) enhances dendritic cell vaccination in experimental melanoma. Oncoimmunology. 2021;10(1):1920739.
doi pubmed - Zhao Q, Yang Y, Liang X, Du G, Liu L, Lu L, Dong J, et al. The clinicopathological significance of neurogenesis in breast cancer. BMC Cancer. 2014;14:484.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.