| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 16, Number 1, February 2025, pages 120-130
VEGFA Gene Expression in Breast Cancer Is Associated With Worse Prognosis, but Better Response to Chemotherapy and Immunotherapy
Pia Sharmaa, Kohei Chidaa, b, Rongrong Wua, c, Kaity Tunga, d, Kenichi Hakamadab, Takashi Ishikawac, Kazuaki Takabea, c, d, e, f, g, h, i
aDepartment of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14203, USA
bDepartment of Gastroenterological Surgery, Hirosaki University Graduate School of Medicine, Hirosaki 036-8562, Japan
cDepartment of Breast Surgery and Oncology, Tokyo Medical University, Tokyo 160-8402, Japan
dDepartment of Surgery, Jacobs School of Medicine and Biomedical Sciences, State University of New York, Buffalo, NY 14203, USA
eDepartment of Breast and Thyroid Surgery, Yokohama City University Medical Center, Yokohama, Kanagawa, Japan
fDivision of Digestive and General Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata 951-8520, Japan
gDepartment of Breast Surgery, Fukushima Medical University School of Medicine, Fukushima 960-1295, Japan
hDepartment of Immunology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14203, USA
iCorresponding Author: Kazuaki Takabe, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14203, USA
Manuscript submitted October 23, 2025, accepted January 2, 2025, published online January xx, 2025
Short title: VEGFA, Prognosis and Treatment Response in BC
doi: https://doi.org/10.14740/wjon1993
| Abstract | ▴Top |
Background: Vascular endothelial growth factor-A (VEGFA) is a key inducer of angiogenesis, responsible for generating new blood vessels in the tumor microenvironment (TME) and facilitating metastasis. Notably, Avastin, which targets VEGFA, failed to demonstrate any significant benefit in clinical trials for breast cancer (BC). This study aimed to investigate the clinical relevance of VEGFA gene expression in BC.
Methods: A total of 7,336 BC patients across eight independent cohorts: ISPY2 (GSE173839), Sweden Cancerome Analysis Network-Breast (SCAN-B) (GSE96058), Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), GSE25066, GSE163882, GSE34138, GSE20194, and The Cancer Genome Atlas (TCGA), were analyzed. The calculated median VEGFA expression level was used to stratify these cohorts into high and low groups.
Results: High VEGFA was associated with worse disease-free, disease-specific, and overall survival in the METABRIC cohort, with findings supported by the SCAN-B cohort, which also showed worse overall survival (all P < 0.02). High VEGFA expression was seen in triple-negative breast cancer (TNBC) but not in BC with lymph node metastasis. Additionally, there was a significant correlation between high VEGFA expression and higher silent and non-silent mutations, single-nucleotide variant (SNV) neoantigens, homologous recombination defect, intratumoral heterogeneity, in the TCGA cohort. In the TCGA, METABRIC, and SCAN-B cohorts, high VEGFA BC was also associated with higher cell proliferation: higher Ki67 gene expression, higher Nottingham histological grade, and consistent enrichment of all the Hallmark cell proliferation-related gene sets. Unexpectedly, the angiogenesis gene set was not enriched in any of the cohorts and showed no association with infiltrations of lymphatic or blood vascular endothelial cells besides pericytes. High VEGFA BC had significantly less infiltration of anti-cancer immune cells but higher infiltration of pro-cancer immune cells in TCGA, METABRIC, and SCAN-B cohorts. Interestingly, BC, which had a pathological complete response (pCR) after anthracycline- and taxane-based neoadjuvant therapy, was associated with significantly heightened VEGFA expression in both estrogen receptor (ER)+/human epidermal growth factor receptor 2 (HER2)- and TNBC subtypes in the GSE25066 cohort and after immunotherapy in ER+/ HER2- subtype, but not TNBC in the ISPY2 cohort.
Conclusions: Our research indicates that high VEGFA BC confers high cell proliferation, reduced immune cell infiltration, and poorer survival, but allows better response to anthracycline- and taxane-based chemotherapy, and immunotherapy.
Keywords: VEGFA; Gene expression; Angiogenesis; Chemotherapy; Immunotherapy; Breast cancer
| Introduction | ▴Top |
Breast cancer (BC) is the second leading cause of mortality in women in the USA [1]. Since the mid-2000s, the incidence rates of BC in females have steadily increased by approximately 0.6% per year [1]. In this recent era of BC treatment, a variety of different therapies have emerged, including surgery, chemotherapy, radiation, immunotherapy, hormone therapy, and targeted therapy [2]. A key area of interest in the development of targeted therapy is angiogenesis (the formation of new blood vessels from existing ones), which is a hallmark of cancer and a critical process that can fuel tumor growth and provide a conduit for cancer to metastasize [3-6]. We have previously demonstrated that in BC, intra-tumoral angiogenesis is associated with inflammation, immune reaction, and metastatic recurrence [7]. Numerous studies have focused on identifying and validating molecular biomarkers for their use as prognostic and predictive indicators [8-10].
A well-known biomarker is the vascular endothelial growth factor-A (VEGFA), which is recognized for its capacity to induce angiogenesis and potential to achieve effective treatment [11, 12]. Recently, our understanding of VEGFA has expanded, and we recognize that VEGFA is pivotal in not only regulating blood vessel formation but also in modifying tumor-induced immunosuppression [13]. VEGFA was evaluated as a prognostic marker but was shown to have no predictive value in metastatic colorectal, renal, and non-small-cell lung cancers [14]. Despite its established role, the clinical implications of VEGFA in BC concerning patient outcomes and therapeutic responses have not yet been understood. Therefore, it was in our interest to investigate the clinical relevance of VEGFA gene expression as both a prognostic and predictive biomarker in BC.
Recently, VEGFA has been recognized as a key target for emerging anti-angiogenic therapies [4, 15]. Due to an initial lack of benefit of the anti-angiogenic therapy bevacizumab for BC and a more recent understanding of VEGFA, combining anti-angiogenic therapies such as bevacizumab with immunotherapies has raised great interest [16]. Bevacizumab (Avastin®) is the first anti-angiogenic targeted therapy that was developed. It has great efficacy for treating colorectal and lung cancers [17] but has been withdrawn from the treatment for BC [18]. Thus, there is a gap in existing research pertaining to the clinical implications of VEGFA expression in BC and its interaction with the tumor microenvironment (TME). This study aims to fill this gap by analyzing VEGFA gene expression across multiple large cohorts of BC patients to understand the clinical relevance of VEGFA expression in BC, its impact on patient outcomes, and its potential as a biomarker for therapy response, potentially leading to the development of more personalized treatment strategies and improved outcomes.
| Materials and Methods | ▴Top |
Clinical data acquisition
A total of 7,336 BC patients from eight publicly available independent cohorts were analyzed, ISPY2 (GSE173839, n = 105), Sweden Cancerome Analysis Network - Breast (SCAN-B) (GSE96058, n = 3,069), Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (n = 1,903), The Cancer Genome Atlas (TCGA) (n = 1,073), GSE25066 (n = 508), GSE20194 (n = 278), GSE163882 (n = 222), and GSE34138 (n = 178). The cohorts analyzed in this study were selected based on their robust sample sizes, and association of clinical parameters and full transcriptome including VEGFA gene expression, which are critical for ensuring comprehensive and statistically reliable analyses. TCGA, METABRIC, and SCAN-B are the largest cohorts representing diverse patient demographics (TCGA is from the USA, METABRIC is from the UK and Canada, and SCAN-B is from Sweden) that we were able to access. Cohorts GSE20194, GSE34138, GSE163882, GSE25066, and ISPY2, were chosen for their detailed drug response data associated with full transcriptome, allowing the assessment of VEGFA expression in relation to therapeutic outcomes. High and low VEGFA gene expression groups were divided based on the calculated median. The TCGA cohort data [19] and the METABRIC cohort data [20] were downloaded from cBioportal as described by Gao et al [21]. The SCAN-B cohort data [22] were downloaded using the R package GEOquery from the NCBI Gene Expression Omnibus database. To analyze the relationship between VEGFA gene expression and drug response, the following primary BC cohorts were analyzed: GSE20194, GSE34138, GSE163882, GSE25066, and ISPY2 [23-27]. These cohorts were also downloaded from the NCBI Gene Expression Omnibus database using the R package GEOquery.
The TCGA, METABRIC, SCAN-B and all GEO cohorts used in this study are deidentified and available within the public domain, therefore approval from the Institutional Review Board was waived. The declaration of ethical compliance with human study was not applicable.
Gene set enrichment analysis (GSEA)
Due to its ability to identify biologically meaningful patterns within large genomic datasets, GSEA [28] was used to investigate which hallmark of cancer gene sets from the Molecular Signatures Database (MSigDB) [29] were associated with VEGFA expression within TCGA, METABRIC, and SCAN-B cohorts. The gene sets were enriched to high or low based on median VEGFA expression. Based on the guidelines of the GSEA MSigDB to balance sensitivity and specificity, the statistical significance was defined as a false discovery rate (FDR) of less than 0.25, and the normalized enrichment score (NES) was used to assess the correlation strength.
Other scores
TCGA samples such as homologous recombination deficiency (HRD) score, intratumoral heterogeneity score, insertions and deletions (Indel) neoantigens, single-nucleotide variant (SNV) neoantigens, and silent and non-silent mutations were calculated and reported by Thorsson et al [30]. The xCell computer algorithm was used to estimate the fraction of intratumoral immune and stromal cells [31]. For each cohort, xCell compared 489 gene signatures corresponding to 64 cell types to estimate the immune and stromal cell fraction [31]. The use of TCGA-specific genomic and immune-related scores, such as HRD and immune cell fractions calculated using xCell, was intended to provide a multi-dimensional view of the TME and genetic alterations.
Statistical analysis
This study was analyzed using R software (version 4.3.1). All P values were calculated by a two-sided test, and the statistical significance was set at 0.05. A Log-rank test was used to test the significance of the survival analysis.
| Results | ▴Top |
High VEGFA expression was associated with lower survival in comparison to the low VEGFA expression group
Given that VEGFA is an established angiogenesis factor, and angiogenesis is associated with metastasis, it was our interest to analyze the relationship between VEGFA expression and patient survival firstly. We found that the VEGFA high expression group showed significantly lower disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in the METABRIC cohort, as well as significantly lower OS in the SCAN-B cohort (all P < 0.02) (Fig. 1). There were no significant survival differences by VEGFA expression in the TCGA cohort, likely due to its smaller sample size. The overall analysis consistently indicated that patients with high VEGFA expression were associated with a worse survival outcome compared to those with low VEGFA expression. This finding aligns with the role of VEGFA in promoting angiogenesis and facilitating metastatic potential.
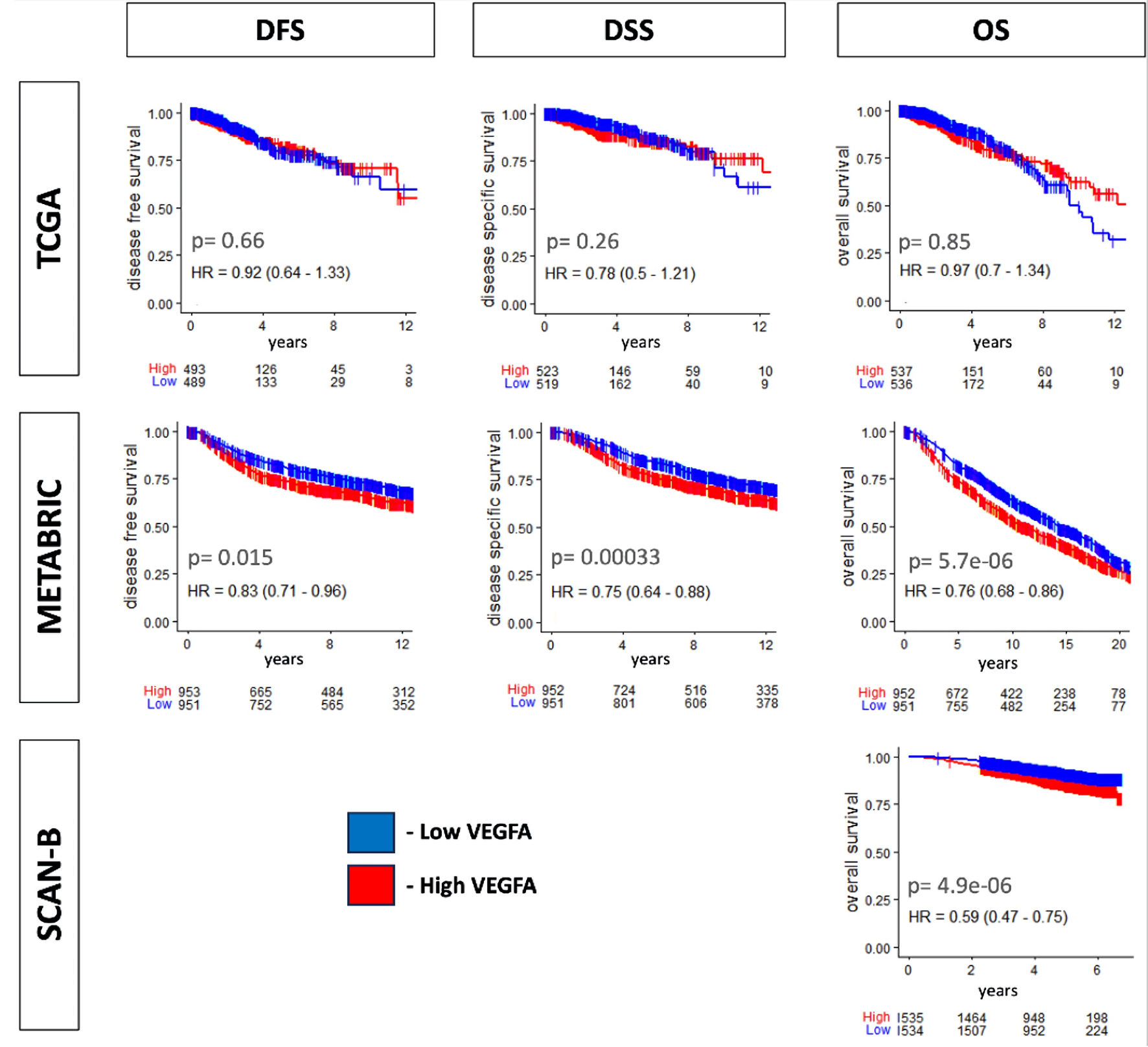 Click for large image | Figure 1. Survival relevance for VEGFA expression. Kaplan-Meier curve with log-rank P value of disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in TCGA and METABRIC and OS in SCAN-B. The median value was used as a cutoff for two VEGFA expression groups, low (blue) and high (red). VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas. |
VEGFA expression was associated with triple-negative breast cancer (TNBC), but not with American Joint Committee on Cancer (AJCC) staging
The finding that high VEGFA expression was associated with a worse clinical prognosis prompted further analysis of clinical parameters, such as the subtype and AJCC staging. High VEGFA expression was found to have a significant association with the TNBC subtype in TCGA, METABRIC, and SCAN-B cohorts (all P < 0.001) (Fig. 2). As TNBC is the more aggressive BC subtype, these results suggest that high VEGFA expression is associated with more aggressive BC. Interestingly, BC with higher VEGFA expression had lower lymph node metastasis in METABRIC and SCAN-B cohorts (P = 0.001 and P = 0.004, respectively). Furthermore, there was no consistent association with distant metastasis.
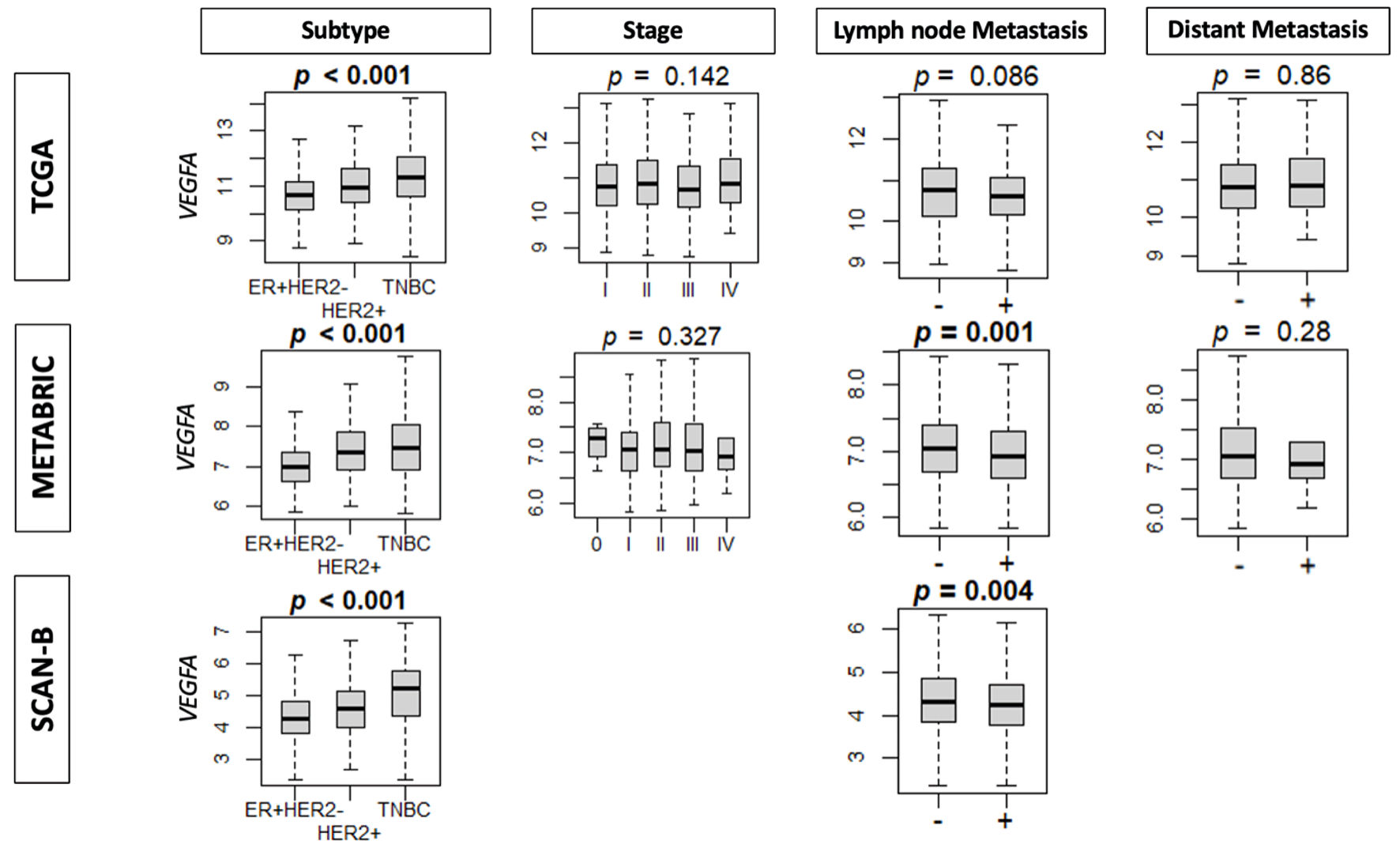 Click for large image | Figure 2. The association between VEGFA expression and clinical parameters. Boxplots of clinical factors: subtype, stage, lymph node metastasis, and distant metastasis in TCGA, METABRIC and SCAN-B cohorts by VEGFA expression. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas. |
VEGFA was consistently associated with cell proliferation and aggressive features of cancer
Cell proliferation is also a key feature of cancer aggression, so it was of interest to investigate the association of VEGFA with mutation rate using scores pre-calculated by Thorsson et al [30] and with cell proliferation. We found that VEGFA was significantly associated with a higher Nottingham histological grade, and more expression of the proliferative marker (Ki67 gene) in TCGA, METABRIC, and SCAN-B cohorts (all P < 0.001) (Fig. 3a). VEGFA also consistently enriched all the Hallmark cell proliferation-related gene sets including MYC targets v1 and v2, E2F targets, G2M checkpoint, mitotic spindle, and mTorC1 signaling in TCGA, METABRIC, and SCAN-B cohorts (Fig. 3b). Additional analyses in the TCGA cohort revealed that high VEGFA expression was linked to higher silent and non-silent mutation rates, HRD, intratumoral heterogeneity, and SNV neoantigens (Fig. 3c).
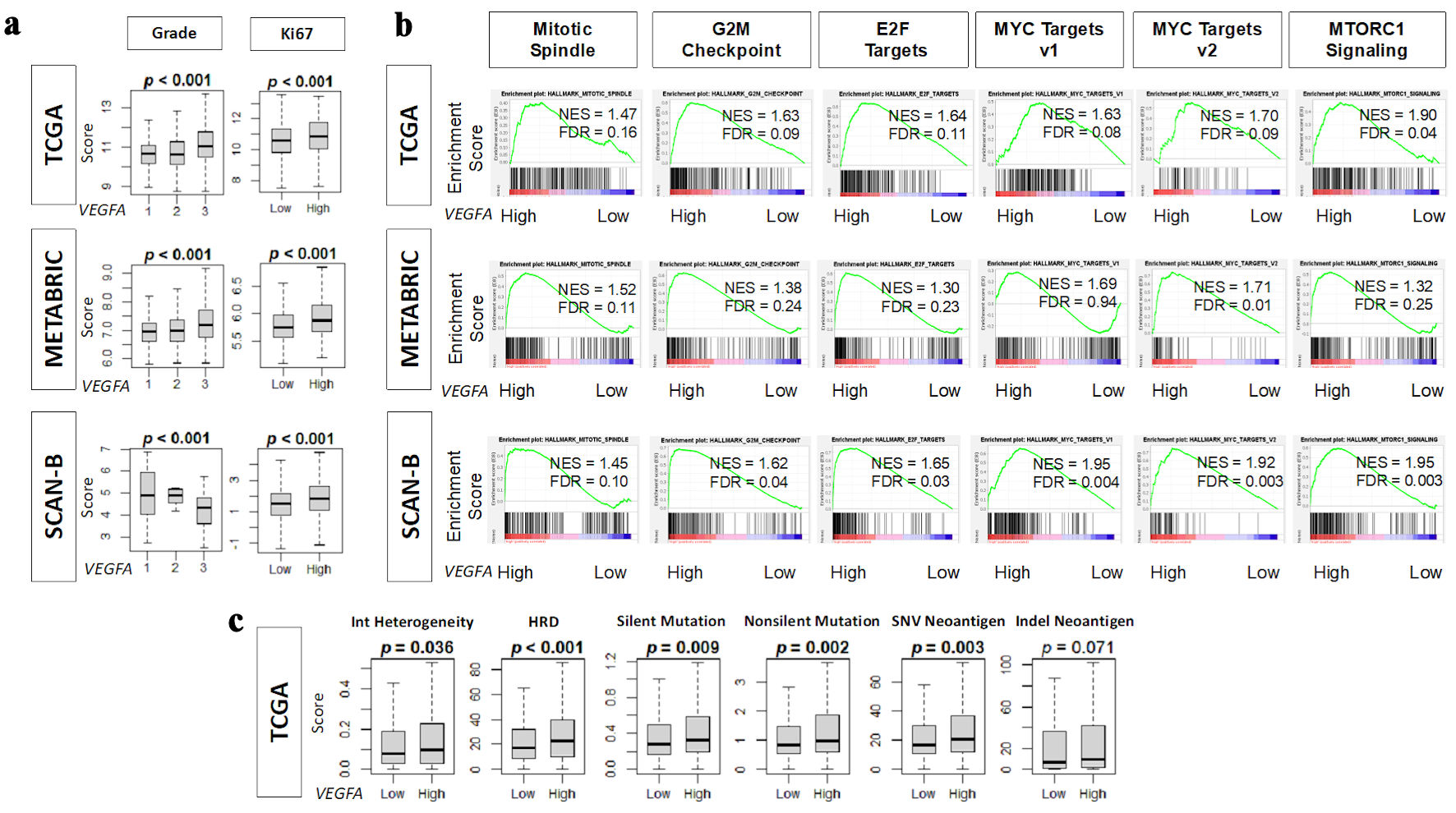 Click for large image | Figure 3. Association of VEGFA with pathological grade, mutation rates, neoantigens, and cell proliferation-related gene sets. (a) Boxplots of pathological grade and Ki67 gene (MKI67) expression in TCGA, METABRIC, and SCAN-B cohorts. (b) Enrichment score plots of cell proliferation-related gene sets: mitotic spindle, G2M checkpoint, E2F targets, MYC targets v1 and v2, and MTORC1 signaling in TCGA, METABRIC, and SCAN-B cohorts by GSEA using NES (normalized enrichment score) and FDR (false discovery rate). As recommended by GSEA software, FDR < 0.25 defined statistical significance. (c) Boxplots of homologous recombination defects (HRD), intratumor heterogeneity, and the mutation-related scores: silent and non-silent mutation rate, single nucleotide variation (SNV) and indel neoantigens in TCGA cohort. High and low VEGFA expression groups were determined by median cutoff. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas; GSEA: gene set enrichment analysis; ITH: intratumor heterogeneity. |
VEGFA was associated with an immunosuppressive TME
To understand how VEGFA might influence the TME, we investigated the association of VEGFA expression with immune cell infiltration within the TME. It was seen that VEGFA was associated with low infiltration of anti-cancer immune cells such as CD8+ T cells, central memory cytotoxic T cells (CD8+ Tcm), CD4+ T cells, CD4+ naive T cells, dendric cells, B cells, and naive B cells, as well as a high infiltration of immune cells, such as type 1 helper T cells (Th1 cells), regulatory T cells, type 2 helper T cells, and plasma cells (Fig. 4).
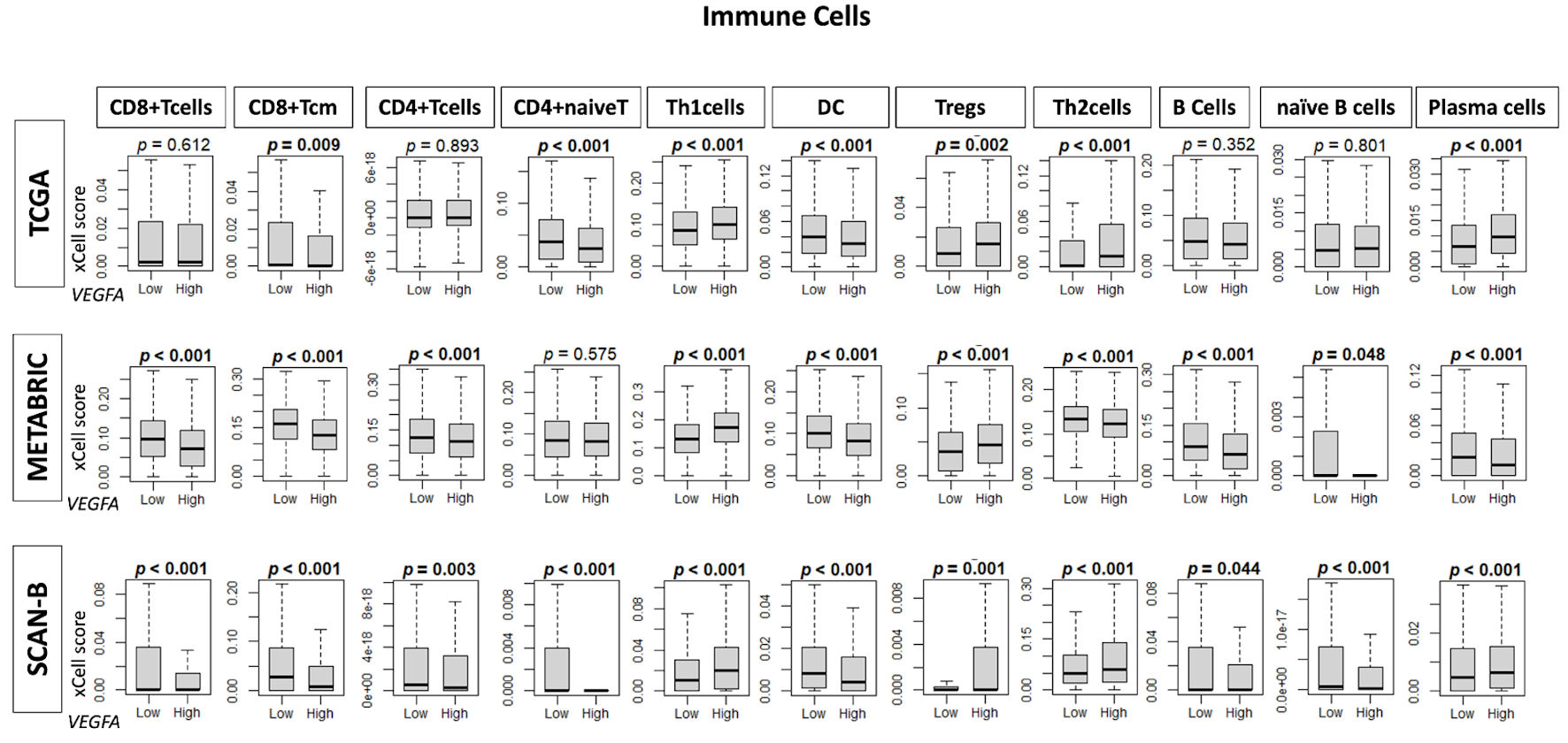 Click for large image | Figure 4. Infiltration fractions of immune cells in the tumor microenvironment by VEGFA expression. Box plots show infiltration fractions for immune cells: CD8+ T cells, CD8+ Tcm cells, CD4+ T cells, CD4+ naive T cells, helper T type 1 (Th1 cells), dendric cells (DC), regulatory T cells (Tregs), helper T type 2 (Th2) cells, B cells, naive B cells, and plasma cells in TCGA, METABRIC, and SCAN-B cohorts by low and high VEGFA expression groups determined by median cutoff. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas. |
VEGFA expression correlated with angiogenesis-related data but not angiogenesis
Since VEGFA is primarily known as an angiogenesis factor, we were particularly interested in investigating the relationship between VEGFA and angiogenesis. There was an association of VEGFA with high infiltration of pericytes, but surprisingly, there was no significant association with the angiogenesis gene set or infiltration of endothelial cells, microvascular endothelial (MVE) cells, or lymphatic endothelial cells (LECs) (Fig. 5).
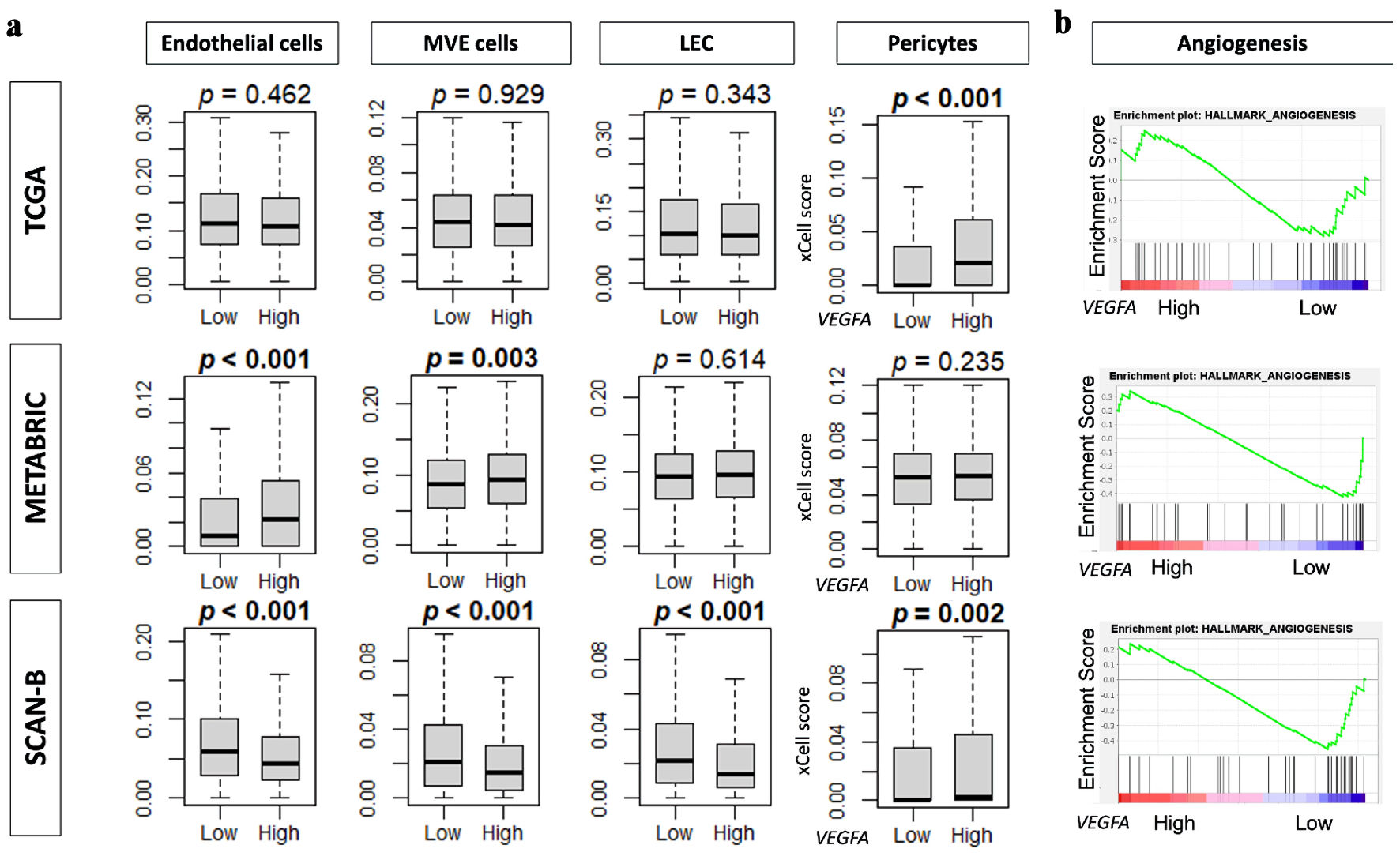 Click for large image | Figure 5. VEGFA association with angiogenesis and angiogenesis-related data. (a) Boxplots of infiltration fractions for angiogenesis-related cells: pericytes, endothelial cells, microvascular endothelial (MVE) cells, and lymphatic endothelial cells (LECs) in in TCGA, METABRIC, and SCAN-B cohorts. (b) Enrichment score plots of angiogenesis gene set by VEGFA high and low groups by GSEA in TCGA, METABRIC, and SCAN-B cohorts. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas. |
Patients with high VEGFA were shown to have a better response to anthracycline- and taxane-based chemotherapy as well as immunotherapy
Due to VEGFA being consistently associated with cancer aggressiveness, it was in our interest to explore the relationship between VEGFA expression and treatment response. Patients with high VEGFA expression showed significantly higher pathological complete response (pCR) with durvalumab/olaparib immunotherapy treatment in comparison to patients with low VEGFA expression in estrogen receptor (ER) positive/human epidermal growth factor receptor 2 (HER2) negative (ER+/HER2-) BC in the ISPY2 cohort. This immunotherapy treatment was also shown to have significantly better results in comparison to the standard chemotherapy group (Fig. 6a). In the GSE25066 cohort, patients with high VEGFA were also shown to have significantly higher pCR with anthracycline- and taxane-based chemotherapy for ER+/HER2- and TNBC, which was supported by the data trends in the GSE20194, GSE163882, and GSE34138 cohorts (Fig. 6b). To ensure that these results were not confounded, all cohorts used were neoadjuvant cohorts. Therapy-specific cohorts such as ISPY2 and GSE25066, were analyzed separately to account for treatment heterogeneity.
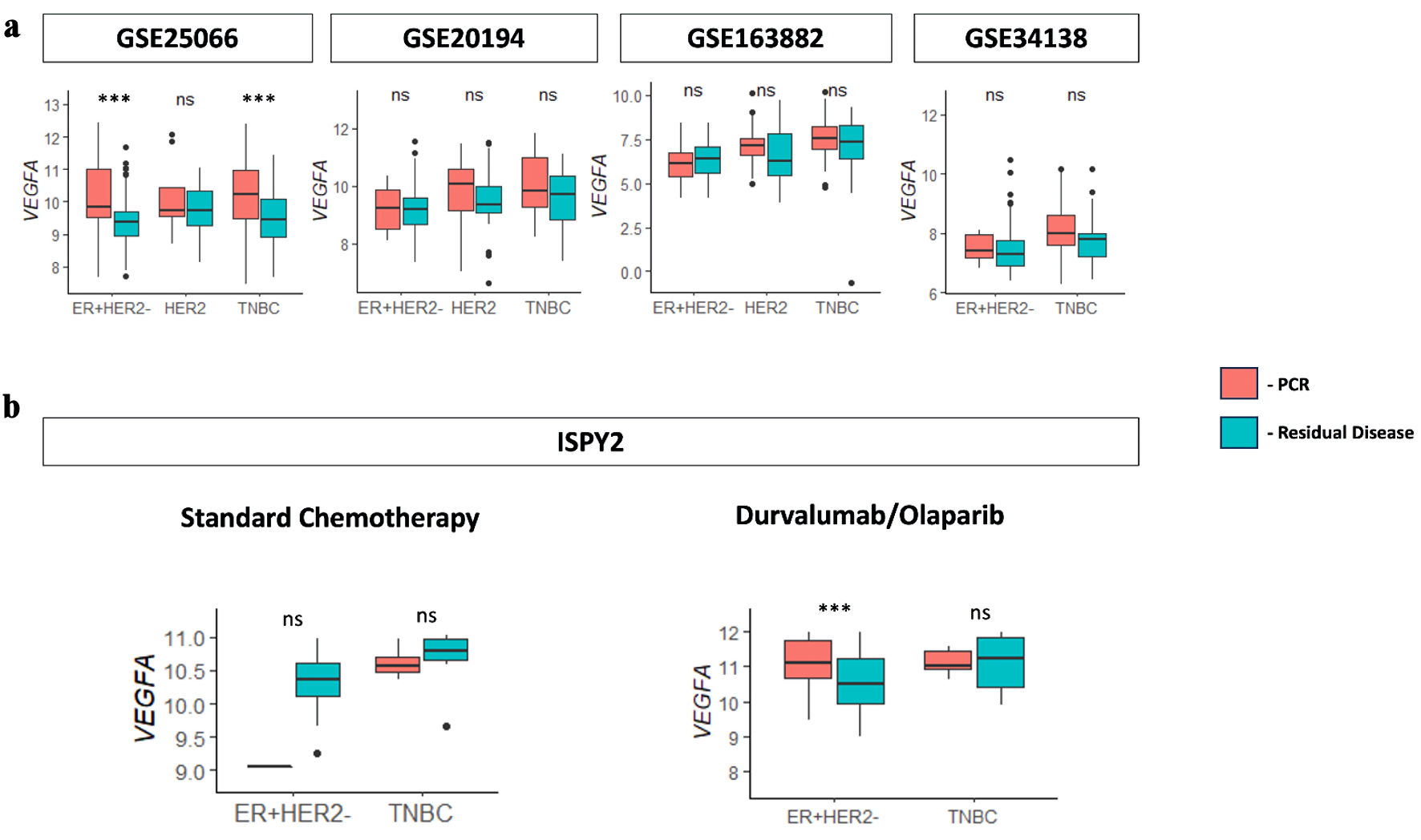 Click for large image | Figure 6. VEGFA association with neoadjuvant therapy patients with pCR. (a) Boxplots for VEGFA expression versus pathological complete response (pCR) and residual disease (RD) response for the immunotherapy treatment (durvalumab/olaparib) group and standard chemotherapy group determined from the ISPY2 cohort. (b) Boxplots for VEGFA expression versus pCR and RD response for anthracycline- and taxane-based chemotherapy in neoadjuvant cohorts GSE25066, GSE20194, GSE163882, and GSE34138. VEGFA: vascular endothelial growth factor-A; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer; NS: not significant. |
| Discussion | ▴Top |
The clinical relevance of VEGFA gene expression was investigated to evaluate its potential as both a prognostic and predictive biomarker in BC. High VEGFA expression was associated with worse OS. Interestingly, VEGFA expression was elevated in TNBC but not in BC with lymph node metastasis. There is also an association of VEGFA expression to increased cell proliferation, as evidenced by elevated Ki67 gene expression, higher Nottingham histological grade, and the enrichment of all the Hallmark cell proliferation-related gene sets, including MYC targets v1 and v2, E2F targets, G2M checkpoint, mitotic spindle, and mTorC1 signaling. Following this trend, VEGFA expression also correlated with significantly higher SNV neoantigens, silent and non-silent mutations, homologous recombination defect, and intratumoral heterogeneity, in TCGA. In addition, VEGFA was associated with low infiltration of anti-cancer immune cells. Also, the angiogenesis gene set was not enriched, and neither blood nor lymphatic vascular endothelial cells, apart from pericytes, were infiltrated in VEGFA-high BC in any of the cohorts. Furthermore, BC, which achieved pCR after anthracycline- and taxane-based neoadjuvant therapy, was associated with significantly higher VEGFA expression in both ER+/HER2- and TNBC subtypes, as well as after immunotherapy in ER+/HER2- subtype, but not TNBC.
Given the significant treatment complexities that cancer poses, there was considerable interest in a promising anti-VEGFA treatment, aiming to suppress angiogenesis by targeting VEGFA, a key inducer [32]. Around 15 years ago, the first approved anti-VEGFA drug bevacizumab (Avastin®, F. Hoffmann La-Roche AG, Switzerland) entered clinical practice [13]. This anti-VEGFA drug revolutionized treatments in cancer, specifically in metastatic colorectal cancer. But unfortunately, it was shown to be ineffective and withdrawn from the treatment of BC [18]. This inefficiency was determined based on two randomized, double-blind, placebo-controlled phase III trials: AVADO and RIBBON-1. The AVADO trial was a three-arm trial evaluating bevacizumab in combination with docetaxel as first-line therapy for HER2-negative, locally recurrent or metastatic BC [33]. The RIBBON-1 trial evaluated anthracycline- or taxane-based chemotherapy with or without bevacizumab as the first-line treatment of HER2-negative, locally recurrent, or metastatic BC [34]. The degree of improvement in progression-free survival (PFS) seen in the AVADO and RIBBON-1 studies did not match the level of improvement observed for the accelerated approval, so bevacizumab was withdrawn from BC treatment [18].
In our previous study, angiogenesis was seen to be associated with an attenuated TME, aggressive biology, and poorer survival outcomes in gastric cancer patients [3]. We have shown that the angiopoietin pathway that promotes angiogenesis was associated with poor survival in BC patients [35]. In our past studies, we have been proficient in performing single-gene analyses of biomarkers using similar methods [36, 37]. We have repeatedly demonstrated that sphingosine 1-phosphate, generated by sphingosine kinase 1, is exported from cancer cells and promotes angiogenesis and lymphangiogenesis in BC [7, 38-42]. It was in our interest to evaluate VEGFA as an angiogenesis factor in BC due to the variability in anti-VEGFA treatment efficacy by cancer type [13].
Aligning with VEGFA’s role in promoting angiogenesis, high VEGFA expression in BC was found to be associated with poorer OS, as demonstrated in the METABRIC cohort and further validated in the SCAN-B cohort. The worse survival outcomes may be attributed to VEGFA’s contribution to creating a more aggressive TME by facilitating the formation of new blood vessels, therefore supporting tumor growth. The finding that VEGFA expression is higher in TNBC, a particularly aggressive subtype with poor prognosis, supports the idea that VEGFA gene expression is associated with more aggressive forms of BC. In contrast, the lower VEGFA levels observed in BC with lymph node metastasis suggest that the role of VEGFA in metastasis could be more complex, potentially influenced by other molecular pathways or factors in the TME. An interesting piece of information that may warrant more investigation was the lack of enrichment of the angiogenesis gene set in all the cohorts, despite VEGFA being a well-established pro-angiogenic factor. Additionally, VEGFA expression did not correlate with the infiltration of lymphatic or blood vascular endothelial cells, except for pericytes. This discrepancy suggests that VEGFA’s role in BC may extend beyond traditional angiogenesis. VEGFA may influence other aspects of the TME, such as immune modulation or cellular proliferation, which could indirectly affect tumor progression and patient outcomes [43]. Further studies are needed to analyze this relationship and determine whether VEGFA’s impact on angiogenesis is context-dependent or modulated by other signaling pathways.
The association between high VEGFA expression and reduced infiltration of anti-cancer immune cells highlights VEGFA’s potential role in immune evasion [44, 45]. This finding is consistent across multiple cohorts (SCAN-B, METABRIC, and TCGA) and suggests that VEGFA may contribute to creating an immunosuppressive TME. Previous studies have demonstrated that angiogenesis and immunosuppression work together as part of the same tissue repair program, including in cancer [46, 47]. By limiting the presence of cytotoxic T cells and enhancing the infiltration of immunosuppressive cell types, VEGFA may help tumors evade immune surveillance, promoting tumor growth and resistance to certain therapies. Interestingly, despite its association with more aggressive tumor characteristics and poorer survival, high VEGFA expression was linked to a better response to anthracycline- and taxane-based chemotherapy as well as immunotherapy. This finding suggests that VEGFA expression could serve as a predictive biomarker for chemotherapy and immunotherapy response in BC. The enhanced response to chemotherapy may be due to VEGFA-induced changes in the TME, such as increased cell proliferation, which could make tumor cells more vulnerable to DNA-damaging agents like anthracyclines and taxanes [48]. Additionally, the better response to immunotherapy in VEGFA-high tumors may be related to the higher mutation rate observed, potentially leading to more neoantigens that could be targeted by the immune system when adequately stimulated by immunotherapy [49].
Our analysis of VEGFA gene expression in BC across eight independent cohorts, including TCGA, METABRIC, and SCAN-B, reveals several notable findings that align with and diverge from previous research. It has previously been reported that high tumor levels of VEGFA predict a poor response to systemic therapy in advanced BC [50]. In contrast, our findings suggest that VEGFA-high tumors had a better response to anthracycline- and taxane-based neoadjuvant therapy and immunotherapy when compared to VEGFA-low tumors. While VEGFA-high tumors may have responded better to systemic therapy, they also were significantly associated with a worse survival rate. This supports the findings of various studies. After bevacizumab failed to show benefit for BC, it was suggested that anti-angiogenic therapies, in combination with immunotherapy, may have successful outcomes in VEGFA-high patients [51]. Recent studies, including those presented at the 2024 San Antonio Breast Cancer Symposium (SABCS), have highlighted innovative approaches to targeting both angiogenesis and immune suppression in BC, using VEGF inhibitors in combination with immune checkpoint inhibitors (ICIs). These novel strategies, such as bispecific VEGFA and ICI therapies, represent an emerging area of research that aims to address the complex relationship between angiogenesis and immune evasion in the TME. This was supported by our studies’ data, which showed that VEGFA-high tumors had a better response to the combination immunotherapy in the ISPY2 trial. This also aligns with our findings that VEGFA-high tumors exhibit an immunosuppressive microenvironment, which could potentially be reversed by dual angiogenesis and immunotherapy approaches. Combining VEGF inhibitors with ICIs could potentially overcome VEGFA-mediated immune evasion while also leveraging the heightened mutation burden observed in VEGFA-high tumors to enhance immunotherapy efficacy. Future studies should explore these combination regimens to determine their efficacy in VEGFA-high BCs. However, preliminary biomarker analyses of bevacizumab trials in various metastatic cancers suggested that VEGFA was a prognostic marker but had no predictive value [52]. In contrast, our study is among the first to demonstrate that VEGFA can be both a prognostic and predictive biomarker for BC. The association of VEGFA with better responses to anthracycline- and taxane-based chemotherapy, as well as immunotherapy, provides a nuanced understanding of its relationship with survival outcomes. We cannot help but speculate that a simplistic approach such as targeting a factor (VEGFA) that is associated with poor prognosis with bevacizumab would also suppress its function to sensitize cancer to the other treatments, which may be one of the reasons that bevacizumab was not effective for BC. Future studies may need to investigate the order and timing of administration of agents with longitudinal assessment of the biomarkers.
Some limitations can be noted in this study. Since it is a retrospective analysis based on patient data from various public cohorts, some detailed clinical information, such as treatment specifics, was unavailable. Despite the promising association of VEGFA with aggressive tumor features and treatment response, the translation of this study’s findings into clinical practice faces several challenges. VEGFA gene expression was assessed in each cohort, thus there is a need for standardized expression thresholds to guide therapeutic decisions. Future investigation of the mechanistic pathways, through which VEGFA contributes to immune evasion and tumor proliferation, is needed to identify potential therapeutic targets. Additionally, exploring combination therapies, such as anti-VEGFA agents with ICIs, is expected to overcome the limitations of anti-angiogenic treatments in BC. These efforts may ultimately improve personalized treatment strategies and outcomes for patients with VEGFA-driven BC.
Conclusions
High VEGFA expression in BC was linked to more aggressive disease characteristics, including increased cell proliferation, reduced immune cell infiltration in the TME, and poorer survival outcomes. However, it was also associated with a better response to anthracycline- and taxane-based chemotherapy, as well as immunotherapy. These findings suggest that VEGFA could serve as a predictive biomarker to guide treatment selection, as well as a prognostic biomarker to identify patients with poor outcomes.
Acknowledgments
KT was supported by US National Institutes of Health grants R37CA248018, R01CA-250412, R01CA251545, R01EB029596, as well as US Department of Defense BCRP grants W81XWH-19-1-0674 and W81XWH-19-1-0111. This research was supported by Rotary Club District 2830 to KC.
Financial Disclosure
No financial disclosures to declare.
Conflict of Interest
The authors have no potential conflict of interest to disclose.
Informed Consent
Not applicable.
Author Contributions
Conceptualization: P. Sharma, K. Chida, and K. Takabe. Data analyses: P. Sharma, R. Wu, and K. Takabe. Writing - original draft preparation: P. Sharma. Writing - review and editing: K. Chida, K. Tung, T. Ishikawa, K. Hakamada, and K. Takabe. Supervision: T. Ishikawa, K. Hakamada, and K. Takabe. Funding acquisition: K. Takabe. All authors have read and agreed to the published version of the manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article. All the dataset used in this study are publicly available.
| References | ▴Top |
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12-49.
doi pubmed - Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535.
doi pubmed - Oshi M, Satyananda V, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, et al. Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients. Am J Cancer Res. 2021;11(4):1659-1671.
pubmed - Cao Y, Langer R, Ferrara N. Targeting angiogenesis in oncology, ophthalmology and beyond. Nat Rev Drug Discov. 2023;22(6):476-495.
doi pubmed - Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674.
doi pubmed - Majidpoor J, Mortezaee K. Steps in metastasis: an updated review. Med Oncol. 2021;38(1):3.
doi pubmed - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, et al. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21(18):6708.
doi pubmed - Brown LF, Berse B, Jackman RW, Tognazzi K, Guidi AJ, Dvorak HF, Senger DR, et al. Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol. 1995;26(1):86-91.
doi pubmed - Yoshiji H, Gomez DE, Shibuya M, Thorgeirsson UP. Expression of vascular endothelial growth factor, its receptor, and other angiogenic factors in human breast cancer. Cancer Res. 1996;56(9):2013-2016.
pubmed - Otrock ZK, Hatoum HA, Musallam KM, Awada AH, Shamseddine AI. Is VEGF a predictive biomarker to anti-angiogenic therapy? Crit Rev Oncol Hematol. 2011;79(2):103-111.
doi pubmed - Melincovici CS, Bosca AB, Susman S, Marginean M, Mihu C, Istrate M, Moldovan IM, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59(2):455-467.
pubmed - Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114-127.
doi pubmed - Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, Chinot OL. Bevacizumab (Avastin(R)) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat Rev. 2020;86:102017.
doi pubmed - Hegde PS, Jubb AM, Chen D, Li NF, Meng YG, Bernaards C, Elliott R, et al. Predictive impact of circulating vascular endothelial growth factor in four phase III trials evaluating bevacizumab. Clin Cancer Res. 2013;19(4):929-937.
doi pubmed - Niu G, Chen X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr Drug Targets. 2010;11(8):1000-1017.
doi pubmed - Bourhis M, Palle J, Galy-Fauroux I, Terme M. Direct and indirect modulation of T cells by VEGF-a counteracted by anti-angiogenic treatment. Front Immunol. 2021;12:616837.
doi pubmed - Keating GM. Bevacizumab: a review of its use in advanced cancer. Drugs. 2014;74(16):1891-1925.
doi pubmed - Sasich LD, Sukkari SR. The US FDAs withdrawal of the breast cancer indication for Avastin (bevacizumab). Saudi Pharm J. 2012;20(4):381-385.
doi pubmed - Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70.
doi pubmed - Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346-352.
doi pubmed - Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
doi pubmed - Saal LH, Vallon-Christersson J, Hakkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, et al. The Sweden Cancerome Analysis Network - Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7(1):20.
doi pubmed - Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28(8):827-838.
doi pubmed - Kersten K, Coffelt SB, Hoogstraat M, Verstegen NJM, Vrijland K, Ciampricotti M, Doornebal CW, et al. Mammary tumor-derived CCL2 enhances pro-metastatic systemic inflammation through upregulation of IL1beta in tumor-associated macrophages. Oncoimmunology. 2017;6(8):e1334744.
doi pubmed - Chen J, Hao L, Qian X, Lin L, Pan Y, Han X. Machine learning models based on immunological genes to predict the response to neoadjuvant therapy in breast cancer patients. Front Immunol. 2022;13:948601.
doi pubmed - Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873-1881.
doi pubmed - Pusztai L, Yau C, Wolf DM, Han HS, Du L, Wallace AM, String-Reasor E, et al. Durvalumab with olaparib and paclitaxel for high-risk HER2-negative stage II/III breast cancer: Results from the adaptively randomized I-SPY2 trial. Cancer Cell. 2021;39(7):989-998.e985.
doi pubmed - Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550.
doi pubmed - Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425.
doi pubmed - Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830.e814.
doi pubmed - Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
doi pubmed - Kristensen TB, Knutsson ML, Wehland M, Laursen BE, Grimm D, Warnke E, Magnusson NE. Anti-vascular endothelial growth factor therapy in breast cancer. Int J Mol Sci. 2014;15(12):23024-23041.
doi pubmed - Miles DW, Chan A, Dirix LY, Cortes J, Pivot X, Tomczak P, Delozier T, et al. Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 2010;28(20):3239-3247.
doi pubmed - Robert NJ, Dieras V, Glaspy J, Brufsky AM, Bondarenko I, Lipatov ON, Perez EA, et al. RIBBON-1: randomized, double-blind, placebo-controlled, phase III trial of chemotherapy with or without bevacizumab for first-line treatment of human epidermal growth factor receptor 2-negative, locally recurrent or metastatic breast cancer. J Clin Oncol. 2011;29(10):1252-1260.
doi pubmed - Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162(1):191-198.
doi pubmed - Satyananda V, Oshi M, Endo I, Takabe K. High BRCA2 gene expression is associated with aggressive and highly proliferative breast cancer. Ann Surg Oncol. 2021;28(12):7356-7365.
doi pubmed - Wu R, Patel A, Tokumaru Y, Asaoka M, Oshi M, Yan L, Ishikawa T, et al. High RAD51 gene expression is associated with aggressive biology and with poor survival in breast cancer. Breast Cancer Res Treat. 2022;193(1):49-63.
doi pubmed - Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55(9):1839-1846.
doi pubmed - Aoyagi T, Nagahashi M, Yamada A, Takabe K. The role of sphingosine-1-phosphate in breast cancer tumor-induced lymphangiogenesis. Lymphat Res Biol. 2012;10(3):97-106.
doi pubmed - Yamada A, Nagahashi M, Aoyagi T, Huang WC, Lima S, Hait NC, Maiti A, et al. ABCC1-exported sphingosine-1-phosphate, produced by sphingosine kinase 1, shortens survival of mice and patients with breast cancer. Mol Cancer Res. 2018;16(6):1059-1070.
doi pubmed - Satyananda V, Oshi M, Tokumaru Y, Maiti A, Hait N, Matsuyama R, Endo I, et al. Sphingosine 1-phosphate (S1P) produced by sphingosine kinase 1 (SphK1) and exported via ABCC1 is related to hepatocellular carcinoma (HCC) progression. Am J Cancer Res. 2021;11(9):4394-4407.
pubmed - Sarkar J, Oshi M, Satyananda V, Chida K, Yan L, Maiti A, Hait N, et al. Spinster homologue 2 expression correlates with improved patient survival in hepatocellular carcinoma despite association with lymph-angiogenesis. World J Oncol. 2024;15(2):181-191.
doi pubmed - Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557-4566.
doi pubmed - Ribatti D. Immunosuppressive effects of vascular endothelial growth factor. Oncol Lett. 2022;24(4):369.
doi pubmed - Yang J, Yan J, Liu B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front Immunol. 2018;9:978.
doi pubmed - Motz GT, Coukos G. The parallel lives of angiogenesis and immunosuppression: cancer and other tales. Nat Rev Immunol. 2011;11(10):702-711.
doi pubmed - Rahma OE, Hodi FS. The Intersection between Tumor Angiogenesis and Immune Suppression. Clin Cancer Res. 2019;25(18):5449-5457.
doi pubmed - de Almeida LC, Calil FA, Machado-Neto JA, Costa-Lotufo LV. DNA damaging agents and DNA repair: From carcinogenesis to cancer therapy. Cancer Genet. 2021;252-253:6-24.
doi pubmed - Xie N, Shen G, Gao W, Huang Z, Huang C, Fu L. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther. 2023;8(1):9.
doi pubmed - Foekens JA, Peters HA, Grebenchtchikov N, Look MP, Meijer-van Gelder ME, Geurts-Moespot A, van der Kwast TH, et al. High tumor levels of vascular endothelial growth factor predict poor response to systemic therapy in advanced breast cancer. Cancer Res. 2001;61(14):5407-5414.
pubmed - Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15(5):325-340.
doi pubmed - Miles DW, de Haas SL, Dirix LY, Romieu G, Chan A, Pivot X, Tomczak P, et al. Biomarker results from the AVADO phase 3 trial of first-line bevacizumab plus docetaxel for HER2-negative metastatic breast cancer. Br J Cancer. 2013;108(5):1052-1060.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.