Figures
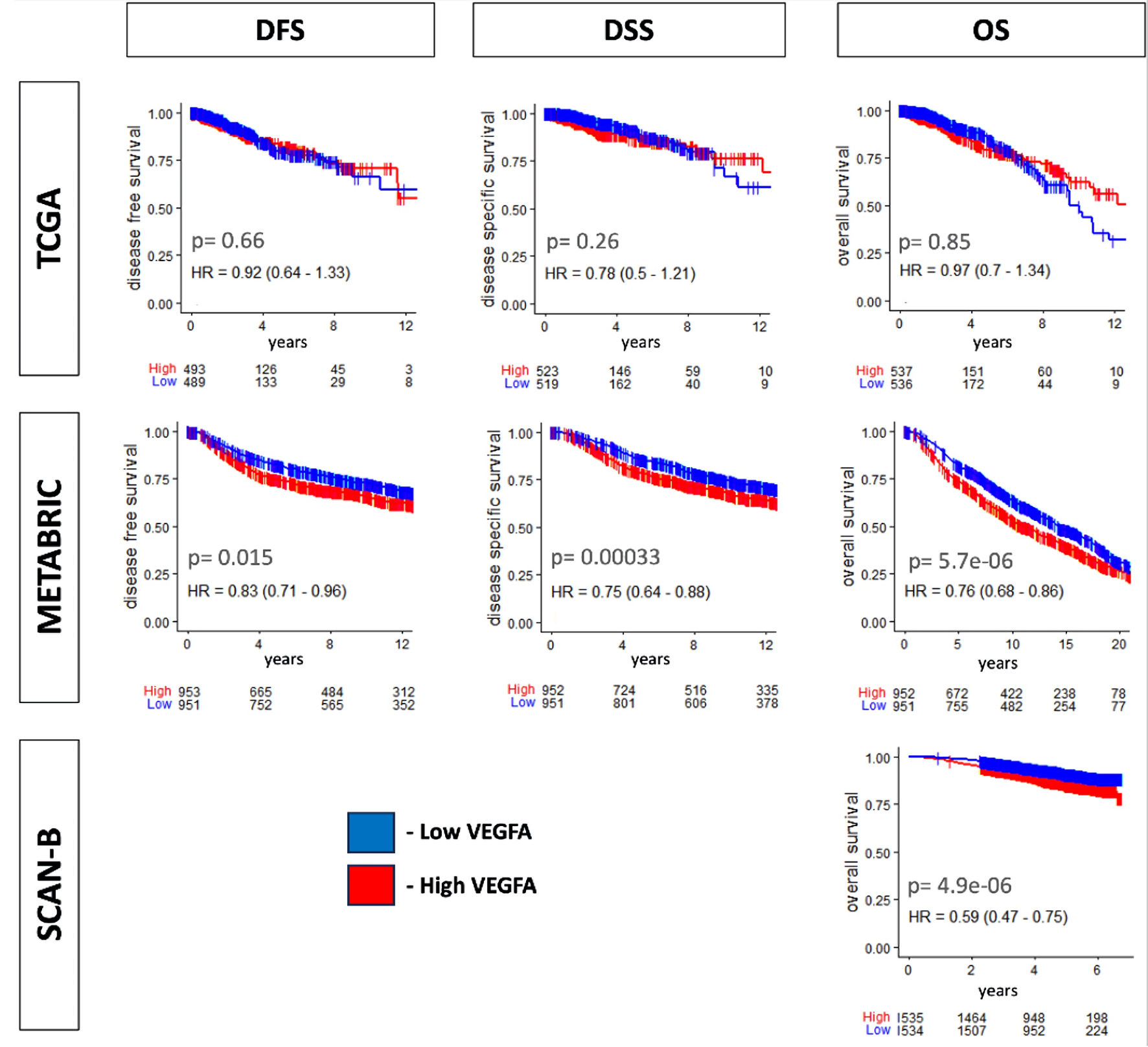
Figure 1. Survival relevance for VEGFA expression. Kaplan-Meier curve with log-rank P value of disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS) in TCGA and METABRIC and OS in SCAN-B. The median value was used as a cutoff for two VEGFA expression groups, low (blue) and high (red). VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas.
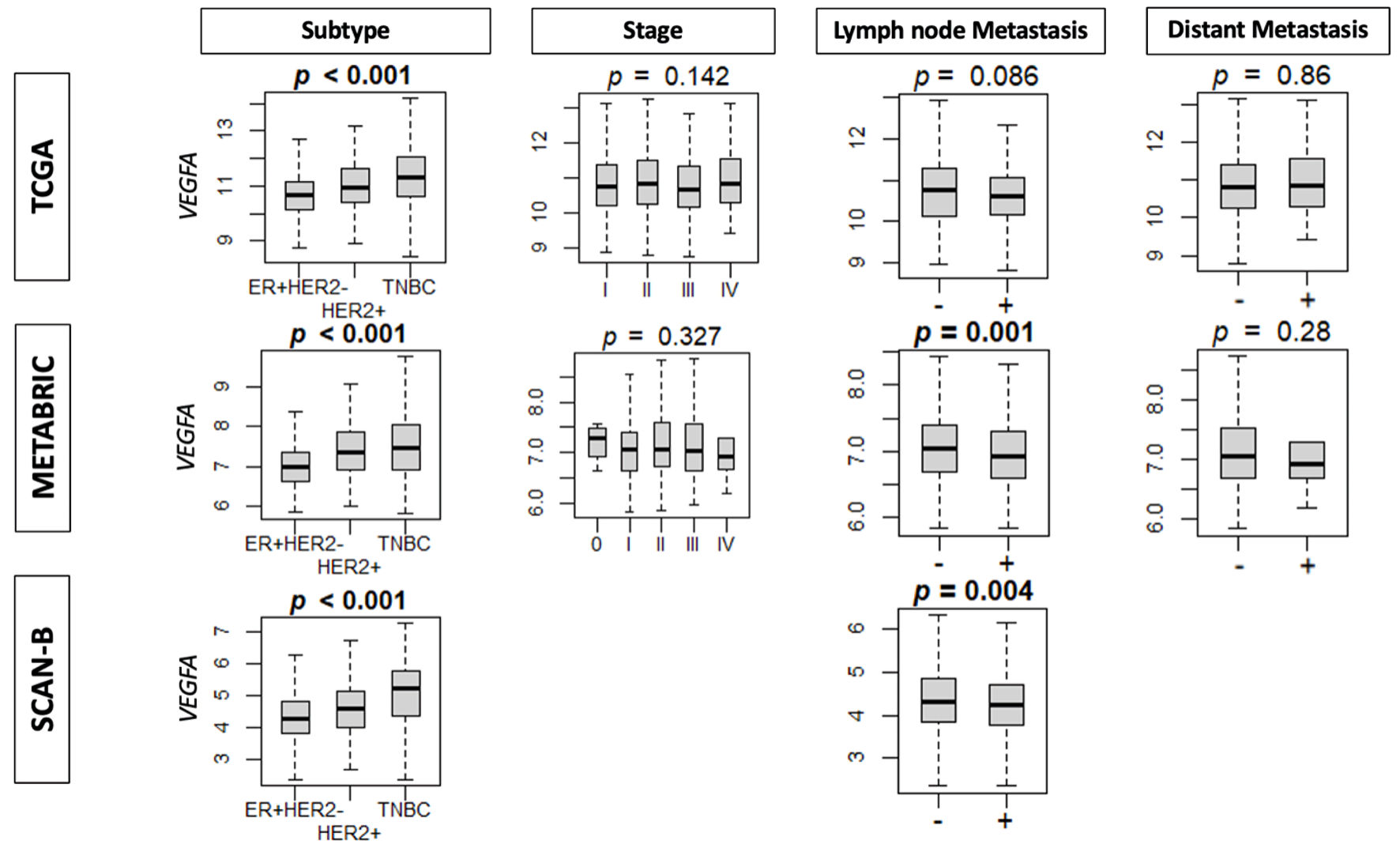
Figure 2. The association between VEGFA expression and clinical parameters. Boxplots of clinical factors: subtype, stage, lymph node metastasis, and distant metastasis in TCGA, METABRIC and SCAN-B cohorts by VEGFA expression. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas.
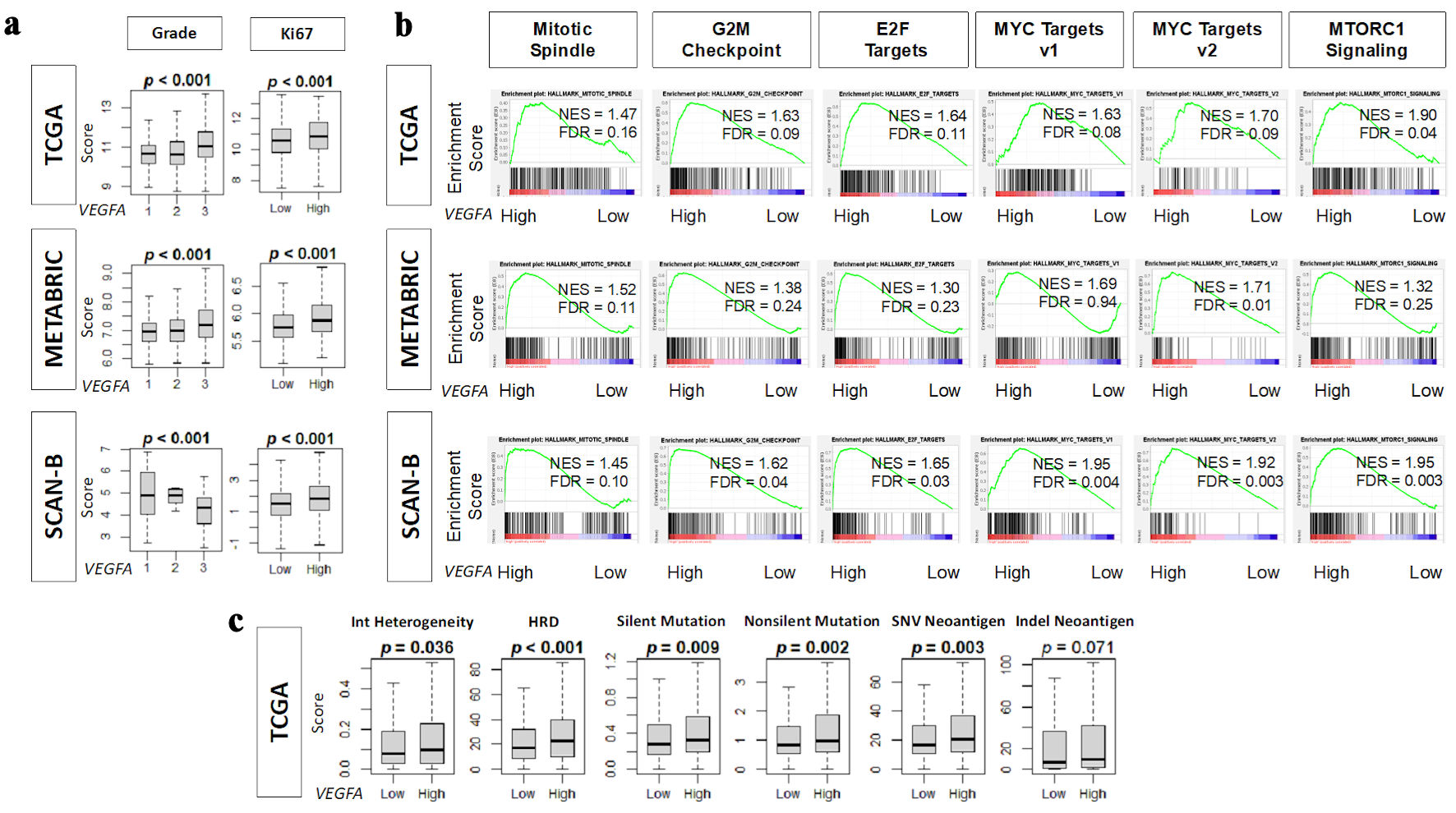
Figure 3. Association of VEGFA with pathological grade, mutation rates, neoantigens, and cell proliferation-related gene sets. (a) Boxplots of pathological grade and Ki67 gene (MKI67) expression in TCGA, METABRIC, and SCAN-B cohorts. (b) Enrichment score plots of cell proliferation-related gene sets: mitotic spindle, G2M checkpoint, E2F targets, MYC targets v1 and v2, and MTORC1 signaling in TCGA, METABRIC, and SCAN-B cohorts by GSEA using NES (normalized enrichment score) and FDR (false discovery rate). As recommended by GSEA software, FDR < 0.25 defined statistical significance. (c) Boxplots of homologous recombination defects (HRD), intratumor heterogeneity, and the mutation-related scores: silent and non-silent mutation rate, single nucleotide variation (SNV) and indel neoantigens in TCGA cohort. High and low VEGFA expression groups were determined by median cutoff. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas; GSEA: gene set enrichment analysis; ITH: intratumor heterogeneity.
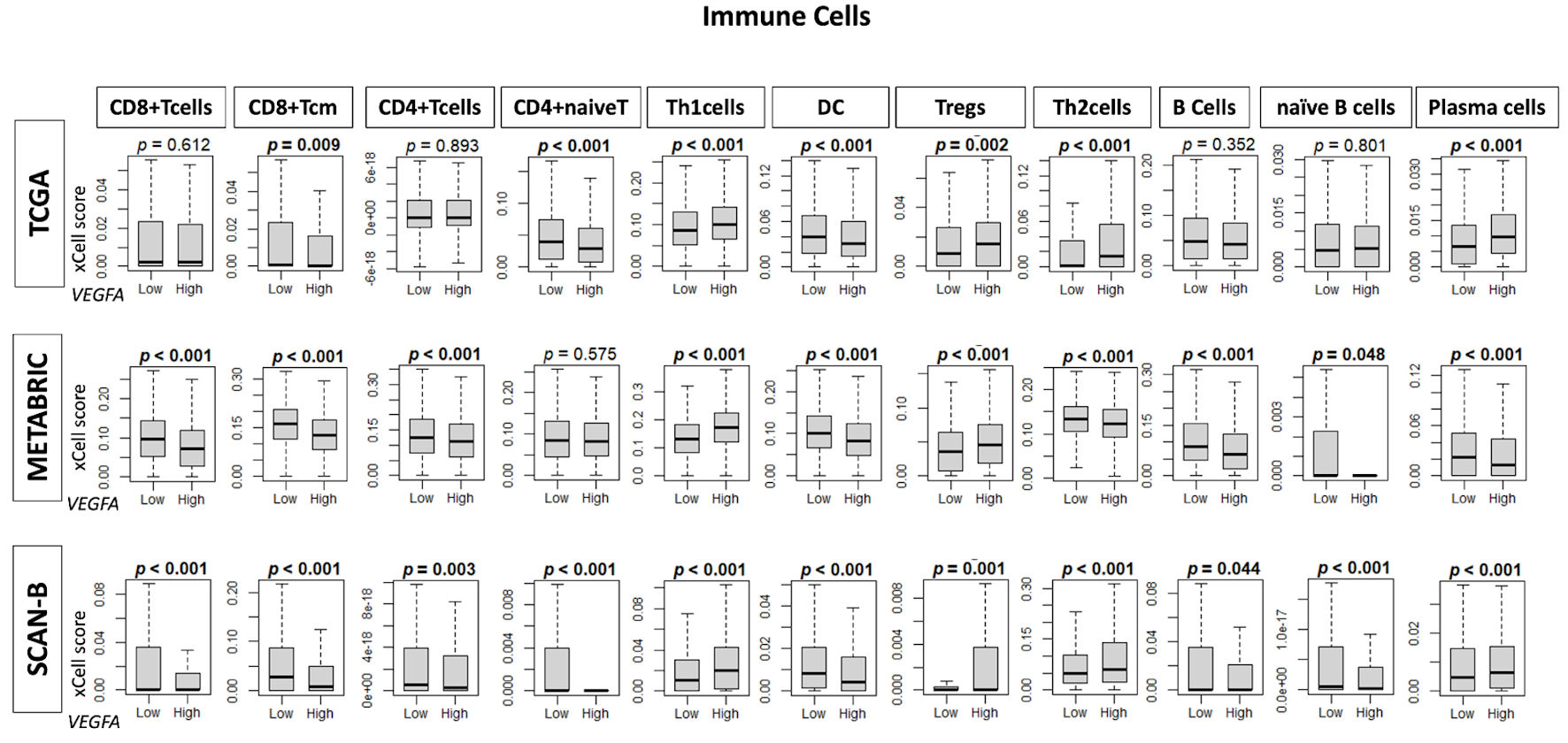
Figure 4. Infiltration fractions of immune cells in the tumor microenvironment by VEGFA expression. Box plots show infiltration fractions for immune cells: CD8+ T cells, CD8+ Tcm cells, CD4+ T cells, CD4+ naive T cells, helper T type 1 (Th1 cells), dendric cells (DC), regulatory T cells (Tregs), helper T type 2 (Th2) cells, B cells, naive B cells, and plasma cells in TCGA, METABRIC, and SCAN-B cohorts by low and high VEGFA expression groups determined by median cutoff. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas.
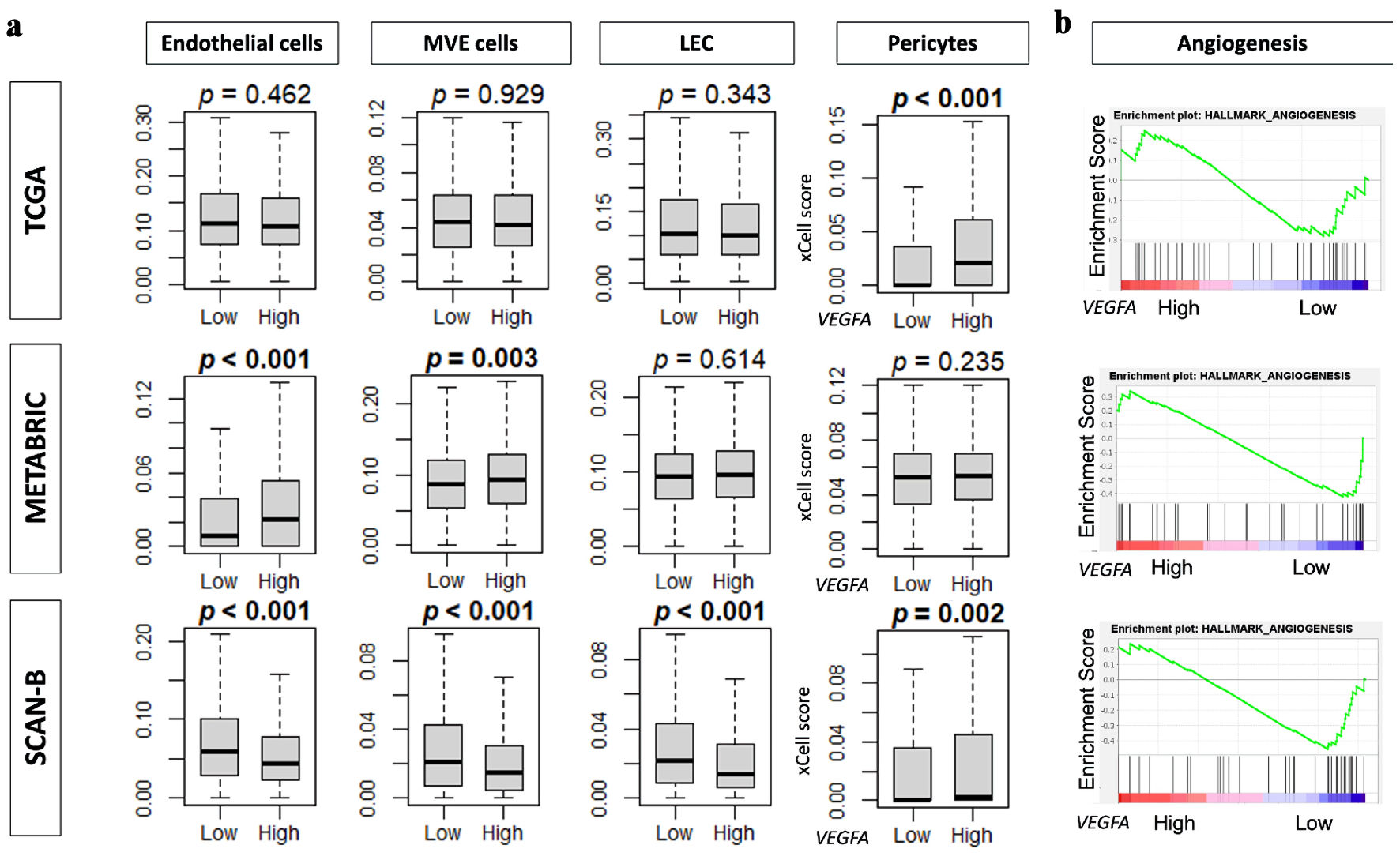
Figure 5. VEGFA association with angiogenesis and angiogenesis-related data. (a) Boxplots of infiltration fractions for angiogenesis-related cells: pericytes, endothelial cells, microvascular endothelial (MVE) cells, and lymphatic endothelial cells (LECs) in in TCGA, METABRIC, and SCAN-B cohorts. (b) Enrichment score plots of angiogenesis gene set by VEGFA high and low groups by GSEA in TCGA, METABRIC, and SCAN-B cohorts. VEGFA: vascular endothelial growth factor-A; SCAN-B: Sweden Cancerome Analysis Network-Breast; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; TCGA: The Cancer Genome Atlas.
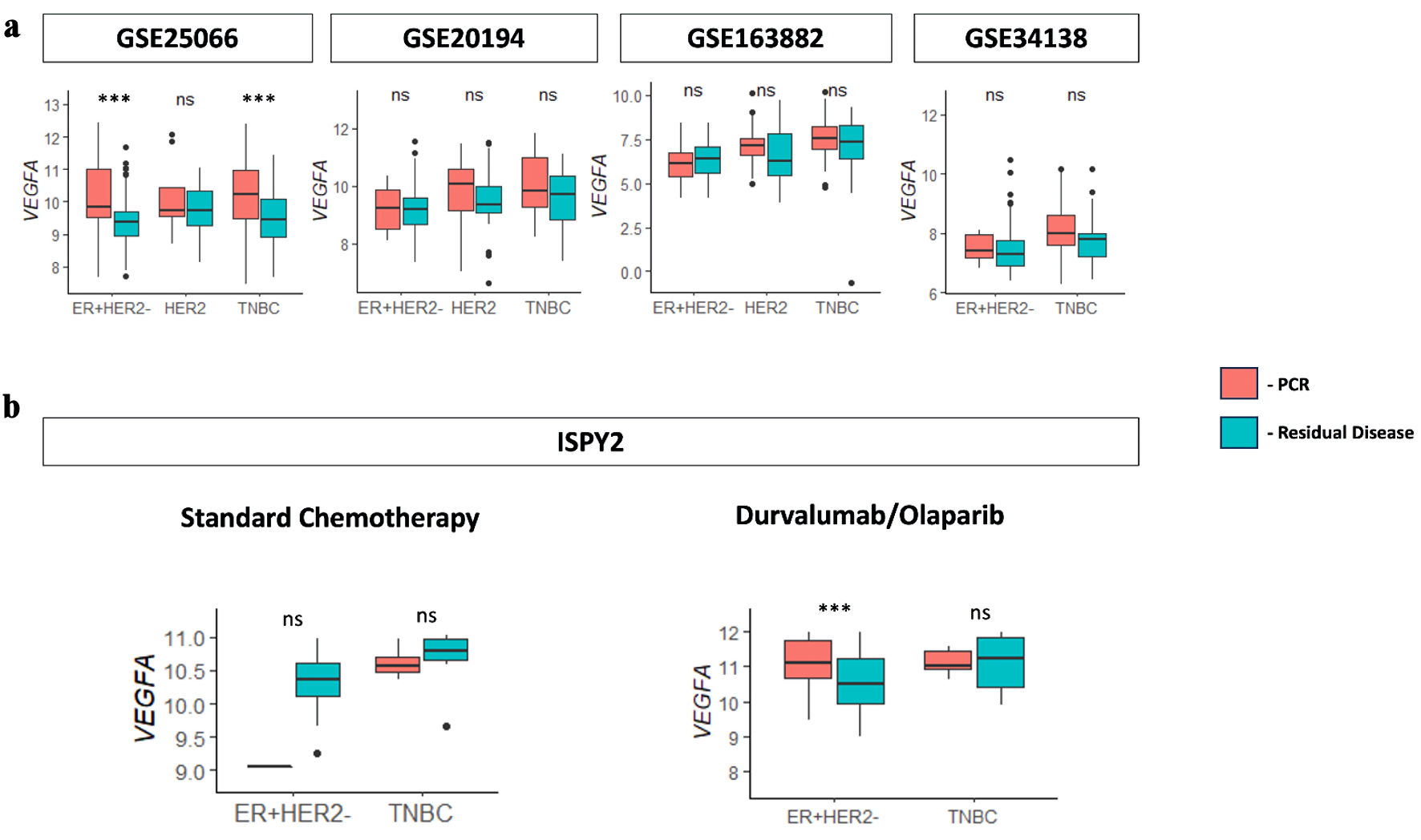
Figure 6. VEGFA association with neoadjuvant therapy patients with pCR. (a) Boxplots for VEGFA expression versus pathological complete response (pCR) and residual disease (RD) response for the immunotherapy treatment (durvalumab/olaparib) group and standard chemotherapy group determined from the ISPY2 cohort. (b) Boxplots for VEGFA expression versus pCR and RD response for anthracycline- and taxane-based chemotherapy in neoadjuvant cohorts GSE25066, GSE20194, GSE163882, and GSE34138. VEGFA: vascular endothelial growth factor-A; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; TNBC: triple-negative breast cancer; NS: not significant.





