| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 16, Number 1, February 2025, pages 131-141
Correlation Between PIK3R1 Expression and Cell Growth in Human Breast Cancer Cell Line BT-474 and Clinical Outcomes
Yi-Fang Tsaia, b, c, Jiun-I Laia, c, d, Chun-Yu Liua, c, d, Chieh-Ning Hsia, b, Chih-Yi Hsuc, e, Chi-Cheng Huanga, b, f, Chin-Jung Fenga, c, g, Yen-Shu Lina, b, c, Ta-Chung Chaoa, c, h, Jen-Hwey Chiua, b, i, k, Ling-Ming Tsenga, b, c, j
aComprehensive Breast Health Center, Department of Surgery, Taipei Veterans General Hospital, Taipei 112201, Taiwan
bDivision of Breast Surgery, Department of Surgery, Taipei Veterans General Hospital, Taipei 112201, Taiwan
cFaculty of Medicine, School of Medicine, National Yang Ming Chiao Tung University, Taipei 112201, Taiwan
dDivision of Medical Oncology, Department of Oncology, Taipei Veterans General Hospital, Taipei 112201, Taiwan
eDepartment of Pathology and Laboratory Medicine, Taipei Veterans General Hospital, Taipei 112201, Taiwan
fInstitute of Epidemiology and Preventive Medicine, College of Public Health, National Taiwan University, Taipei 100233, Taiwan
gDivision of Plastic Surgery, Department of Surgery, Taipei Veterans General Hospital, Taipei 112201, Taiwan
hDivision of Cancer Prevention, Department of Oncology, Taipei Veterans General Hospital, Taipei 112201, Taiwan
iDivision of General Surgery, Department of Surgery, Cheng-Hsin General Hospital, Taipei 112401, Taiwan
jThese authors contributed equally to this work.
kCorresponding Author: Jen-Hwey Chiu, Comprehensive Breast Health Center, Department of Surgery, Taipei Veterans General Hospital, Taipei 112201, Taiwan
Manuscript submitted October 21, 2024, accepted December 16, 2024, published online January 10, 2025
Short title: Role of PIK3R1 in Breast Cancer
doi: https://doi.org/10.14740/wjon1986
| Abstract | ▴Top |
Background: While mutations in the PIK3CA gene play important roles in human breast carcinogenesis, PIK3R1 gene alterations are recognized as actionable mutations for clinical cancer treatment. We aimed to elucidate the role of PIK3R1 in cell proliferation on breast carcinoma and to correlate the PIK3R1 expression with patients’ outcome using human tumor tissue arrays.
Methods: Using human BT-474 (estrogen receptor (ER)+/human epidermal growth factor receptor 2 (HER2)-high) breast carcinoma cell line as in vitro model, the role of PIK3R1 in cell proliferation was elucidated by knock-down of the PIK3R1 gene (ΔPIK3R1) in this cell line. Between January 2000 to December 2015, the records of a cohort of 440 patients in our hospital were retrospectively reviewed, including patients’ survival. The correlations between PIK3R1 expression and patient prognosis, such as overall survival (OS) and disease-free survival (DFS), were elucidated by human breast cancer tumor tissue array immunostaining.
Results: After the PIK3R1 gene was silenced in the BT-474 line, there was an increased cell number and a decrease in the G0G1-fraction, and increased S-fraction and the S+G2M-fraction for the ΔPIK3R1-BT-474 cell line, as compared to their cell wild type (WT) line. Western blot analysis showed that decreased PIK3R1 protein levels were accompanied by an increase of the p-AKT and p-mTOR proteins in the ΔPIK3R1-BT-474 cell line, compared to the equivalent WT line. Using a human tumor tissue array, patients with high-expressed PIK3R1 protein had better outcomes in terms of DFS and OS, compared to those with low-expressed PIK3R1 protein, when breast cancer was at an early stage (stage I/II), but not across all stages of breast cancer in human patients.
Conclusions: We concluded that downregulated PIK3R1 in BT-474 cells resulted in an increased cell growth and upregulated AKT-mTOR signaling. Clinically, the high-expressed PIK3R1 protein in tumors correlates positively with patients’ outcome in stage I and II breast cancer.
Keywords: Breast cancer; Genetic mutation; PIK3R1; Variants; MCF-7
| Introduction | ▴Top |
Female breast carcinoma is a common malignancy worldwide and in Taiwan [1, 2]. Based on protein expression by immunostaining, proliferative capacity in tumors, and gene expression profiling, breast cancers are categorized into four types, namely, human epidermal growth factor receptor 2 (HER2)-enriched, luminal A, luminal B, and basal-like subtypes [3-6]. Molecular subtyping provides optimal treatment strategies that correlate well with clinical outcomes [7, 8].
Precision medicine is known to identify accurately effective therapies for individual cancer patients, which has increased survival rates. This has become possible because of the rapid development of human genome sequencing and the application of large-scale biological databases to the characterization of patients, and the results can then be used to guide clinical practice. Among the many cancer-related genetic alterations, some of them defined as actionable mutations have been identified in melanoma [9] and non-small cell lung cancer [10] patients. These mutations-related malignancies are responsive to a specific target therapy that has a demonstrable clinical benefit [11].
There is consensus that therapies using poly (adenosine diphosphate (ADP)-ribose) polymerase (PARP) inhibitors result in significantly improved survival outcomes for patients with BRCA1/BRCA2 mutation-associated breast carcinoma [12, 13]. Recent evidence suggests an important role for the phosphatidylinositol 3-kinase (PI3K) catalytic subunit, PIK3CA, in human breast cancers [14-16]. Nonetheless, the exact role of PIK3R1 in human breast carcinoma and its clinical impact have not been explored in depth up to now.
PI3K is composed of a 110 kD catalytic subunit (PIK3CA) and a regulatory subunit (85 kD, 55/50 kD) [17]. The PI3K regulatory subunit1 (PIK3R1) gene encodes the 85 kD regulatory alpha subunit of the enzyme. While mutations in the PIK3CA gene are recognized to play important roles in human breast carcinogenesis, mutations in PIK3R1 are historically associated with insulin resistance and the SHORT syndrome [18]. Furthermore, human PIK3R1 mutations are known to disrupt lymphocyte differentiation, which causes PI3Kδ syndrome 2 [19]. Previously, mutations affecting PIK3R1 have been identified as an actionable mutation for prostate and ovarian cancers [20, 21]. In addition, somatic mutations in PIK3CA (44%), PIK3R1 (17%), and PIK3CA + PIK3R1 (9.0%) have been identified in Chinese population who have breast carcinoma [22]. Using 380 hot spot sequencing analyses, 66% of these cancer samples exhibited ≥ one actionable alternation when the ESCAT criteria were used [23, 24]. Although a poor outcome is correlated well with PIK3R1 mutations among patients with receptor (+) breast carcinoma [25], controversy still exits as to whether the various different breast cancer subtype tumors are associated with activating mutations that cause abnormal activity of the PI3K/Akt/mTOR pathway [26, 27]. Since PI3K activates AKT, a kinase of prime importance to cell survival and proliferation during oncogenic process, it was our aim to elucidate the PIK3R1-related cell growth activity in breast cancer cells and then to explore the correlation between PIK3R1 expression and clinical outcomes in patients using a human tumor tissue array.
| Materials and Methods | ▴Top |
BT-474 cell line and reagents
The BT-474 (estrogen receptor (ER)+/HER2-high) cell line has been obtained from the Food Industry Research and Development Institute (Hsinchu, Taiwan, Republic of China) [28]. BT-474 cells were cultured in Hybri-Care medium supplemented with penicillin/streptomycin and 2 mM L-glutamine at 37 °C in a humidified atmosphere containing 5% CO2. The contamination of mycoplasma in this line was excluded in this study.
Gene manipulation of the human breast cancer BT-474 cell line
Following the protocols obtained from the Academia Sinica, Taiwan, lentiviral infection using short hairpin RNA (shRNA) silencing of the PIK3R1 gene (ΔPIK3R1) in the BT-474 cell line was performed. Real-time polymerase chain reaction (PCR) and Western blot analysis were conducted to validate the infection efficiency (Supplementary Material 1, wjon.elmerpub.com).
Cell proliferation assay
Both the wild type (WT) and ΔPIK3R1 line (1 × 104/well) were cultured with low serum medium. After 1 - 4 days, the cells were washed twice with phosphate-buffered saline (PBS, pH 7.4), and then trypsinized using 0.5 mL trypsin-ethylenediamine tetraacetic acid containing 0.05% trypsin and 0.53 mmol/L ethylenediamine tetraacetic acid I 4Na (Gibco/Invitrogen, New York, NY). The cells were then resuspended in fresh culture medium, and the cell count was measured using trypan blue exclusion assay.
Western blotting analysis
Homogenized BT-474 (both the WT and ΔPIK3R1 lines) cells were lysed with a buffer (150 mM KCl, 10 mM Tris pH 7.4, 1% Triton X-100) containing cocktails of phosphatase and protease inhibitors (Complete Mini; Roche, Mannheim, Germany). The protein concentration of these homogenates was quantified using the Bradford’s method [29]. Next, 30 µg of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 10%) and then transferred to a polyvinylidene fluoride (PVDF) membrane, followed by blocking the membrane with bovine serum albumin (5%) and by probing with specific primary antibodies such as PI3KR (p85 alpha, # PA5-29613, ThermoFisher, Waltham, MA), p-Akt (Ser473) (#9271), p-mTOR (Ser2448) (#2971), Akt (#9272), mTOR (#2983) and β-actin (internal control, #3700). The latter four were obtained commercially (Cell Signaling, Beverly, MA). The washed blots were incubated in anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)-linked secondary antibodies (Cell Signaling Technology, Beverly, MA). After 1 h of hybridization, the membranes were washed and developed with an enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech Inc., NJ) and quantified using Multi-Gauge software (Fuji Photo Film Co., Ltd., Tokyo, Japan).
Total RNA extraction and reverse transcription PCR
Total RNA was obtained using the guanidinium thiocyanate method as described previously [30]. (TRI REAGENT, T-9424, Sigma Chem. Co., St. Louis, MO). A cDNA synthesis kit (Invitrogen, CA) was used to obtain the complementary DNA from a total RNA extract. The changes in de novo gene synthesis for each treatment group were evaluated by reverse transcriptase-polymerase chain reaction (RT-PCR). The primers pair for PIK3R1: 5'-TAGCTCGCGCGATCTAGGGGC-3' (sense) and 5'-CGCGATCAATAAAGCTAG-3' (antisense) were commercially available. The relative expression of the mRNA of each gene was normalized against the glyceraldehyde-3-phosphate dehydrogenase (GAPD) mRNA present in the same RNA extract.
Proliferative activity analyzed by flow cytometry
Cell proliferation was evaluated by cell cycle analysis. Briefly, 1 × 106 cells (both WT and ΔPIK3R1) were seeded and then grown for 1 day, followed by changing the medium containing 0.1% fetal bovine serum and cultured for another 2 days. Then the cells were trypsinized, washed, and fixed with ethanol (75%). Following this, the cells were stained using propidium iodide (PI) and for cell cycle analysis by flow cytometry. The results are presented as the various cell cycle fractions in percentage including G0/G1 phase, S phase, and G2/M phase.
Subjects
During the period from January 2000 to December 2015, patients with breast carcinoma underwent pathological diagnosis at Taipei Veterans General Hospital, Taiwan, Republic of China. With the Institutional Review Board (IRB) approval (# 2022-01-016CC), 440 patient records containing diagnosis date, immunohistochemical (IHC) staining results, such as ER, progestin receptor (PR), HER2 status including IHC and fluorescence in situ hybridization (FISH), and patients’ survival (disease-free survival (DFS) and overall survival (OS)), were retrospectively reviewed from the hospital breast cancer database. Written consent from the study subjects was waived by the IRB because all clinical data had been collected during routine clinical care in the hospital, and no direct contact with patients for data collection and analysis was required. The mean follow-up time for each patient was > 60 months. Receptor status was defined as negative when the ER or PR was ≤ 1%, while an ER or PR of > 1% was regarded as positive. The study was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
Immunohistochemistry for PIK3R1 expression
PIK3R1 expression in a tissue array containing the above 440 tumor samples was assessed by IHC staining and evaluated by an expert in pathology (Dr. Hsu, CY). Positivity for PIK3R1 expression was semi-quantified and expressed as 0 (no staining), 1+ (cytoplasmic staining in less than 10% of tumor cells), 2+ (cytoplasmic staining between 10% and 49% of tumor cells), and 3+ (cytoplasmic staining more than 50% of tumor cells).
Statistics analysis
Data are expressed as means ± standard error of the mean (SEM). Statistical significance between two groups were identified by the Mann-Whitney U test or the Student’s t-test. Comparison between two independent variables was determined by two-way analysis of variance (ANOVA) and the Bonferroni post-hoc test. The Kaplan-Meier analysis was performed to estimate patients’ outcomes. A P value less than 0.05 is considered statistically significant compared to control group or WT group.
| Results | ▴Top |
Expression of PIK3R1 in various breast carcinoma cell lines
To elucidate the PIK3R1 protein expression in different subtypes of breast carcinoma cell lines, we measured the expression in the MCF-7 (luminal A), BT-474 (luminal B), SK-BR-3 (HER-2 enriched), MDA-MB-231/468/453 and BT-549 (basal-like) cell lines, which were maintained in our laboratory. Western blot analyses showed diverse PIK3R1expression patterns across the various subtypes of human breast carcinoma cell lines (Supplementary Material 2, wjon.elmerpub.com).
PIK3R1 regulated the cell proliferation of the BT-474 cell line
Then, we silenced the PIK3R1 gene in the BT-474 cell line. Cell proliferation was observed under a light microscope (Fig. 1a), and the quantification of cell number was evaluated by trypan blue exclusion assay. Our results showed that there was an increased cell number for the ΔPIK3R1-BT-474 cell line, as compared to their cell WT lines (two-way ANOVA) (Fig. 1b).
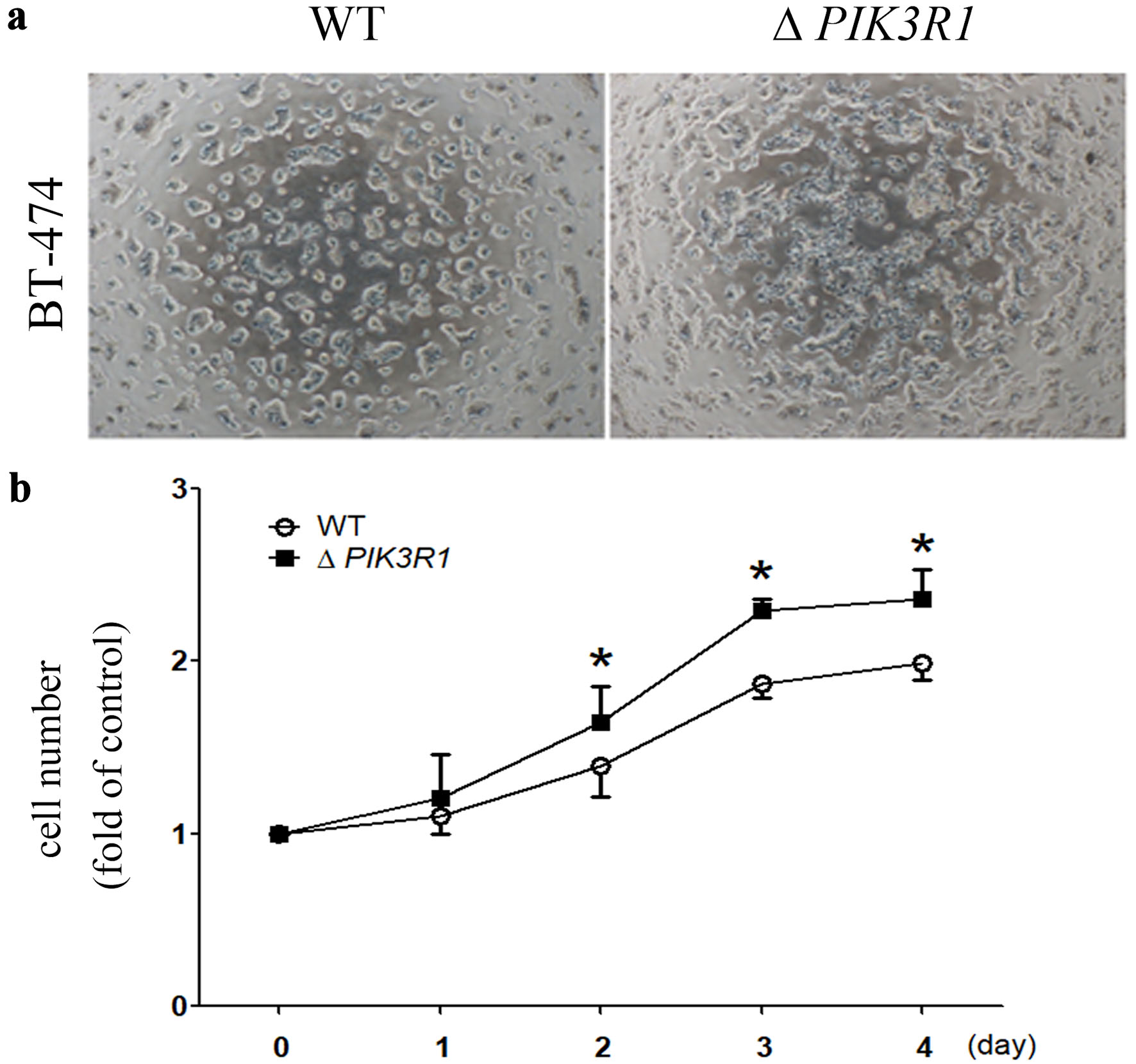 Click for large image | Figure 1. Role of PIK3R1 on cell proliferation in human breast cancer BT-474 cell line. After the PIK3R1 gene was silenced in the BT-474 cell line, the resulting two cell lines (WT and ΔPIK3R1) were cultured and photographed under low power (× 10) using a microscope (a). Cells numbers (WT and ΔPIK3R1) were quantified using the trypan blue exclusion assay (b). Asterisks indicate a P value < 0.05 (two-way ANOVA). WT: wild type; ANOVA: analysis of variance. |
PIK3R1 regulated the cell cycle of the BT-474 cell line
To elucidate how PIK3R1 modulates the cell cycle in the BT-474 cell line, cell cycle analysis (Fig. 2a) using flow cytometry was performed, and the results showed a decrease in the G0G1-fraction, and increased S-fraction and the S + G2M-fraction in ΔPIK3R1-BT-474 cell line compared to WT cell line (Mann-Whitney U test) (Fig. 2b).
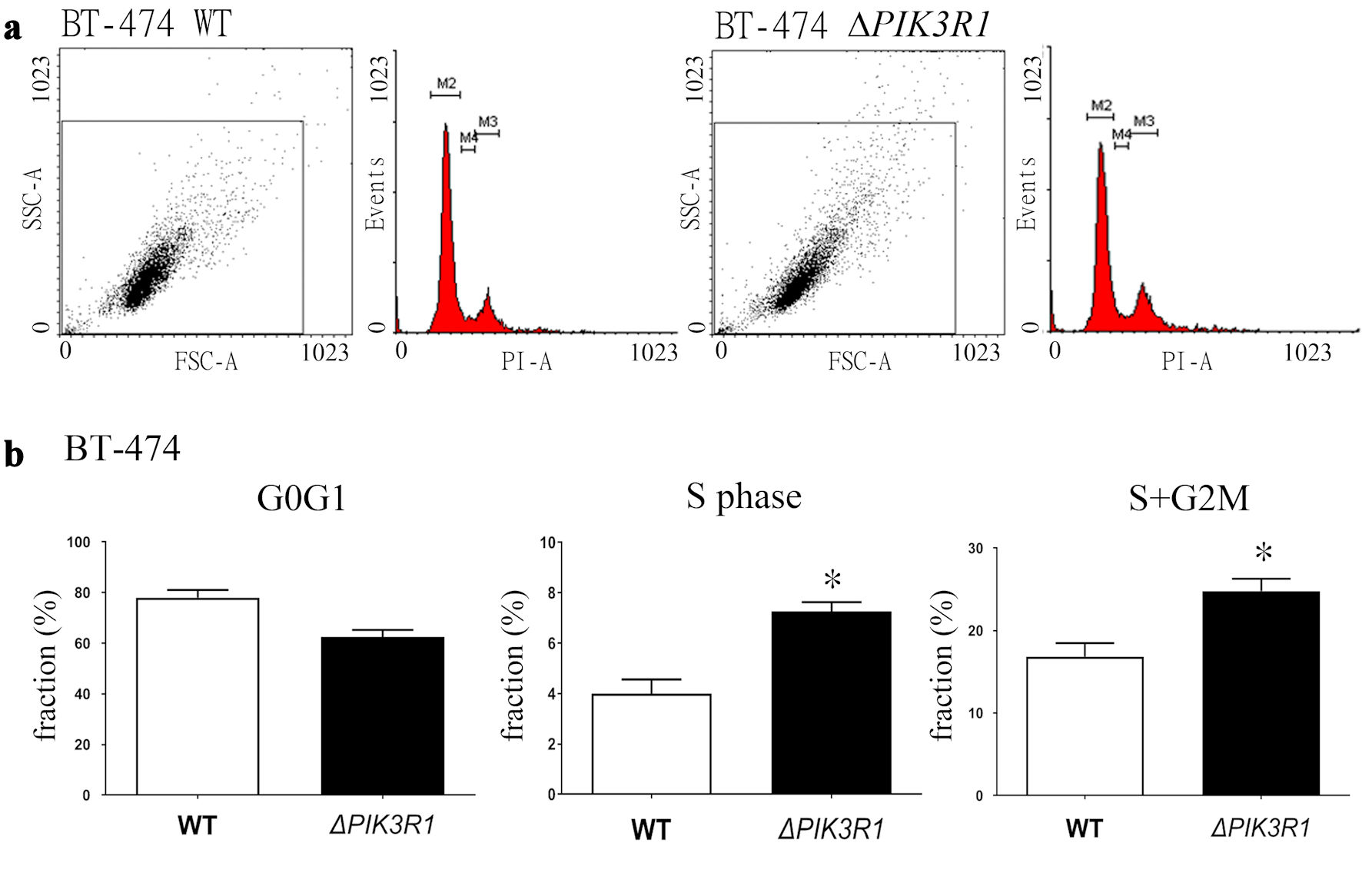 Click for large image | Figure 2. Cell cycle analysis of the ΔPIK3R1 and WT breast cancer BT-474 breast cancer cell lines. (a) For cell cycle analysis, 1 × 106 cells (both WT and ΔPIK3R1) were seeded as described in the Methods. (b) The results are presented and quantified as the various cell cycle fraction (%), namely G0/G1 phase, S phase, and G2/M phase. Asterisks indicate a P value < 0.05 by Mann-Whitney U test. FSC: forward scatter; SSC: side scatter; PI: propidium iodide; WT: wild type. |
PIK3R1 regulated the cell proliferation-related signaling in the BT-474 cell line
After PIK3R1 gene was knocked down in the BT-474 cell line, the PIK3R1 protein was significantly decreased (Fig. 3a, b). This was accompanied by the increase of the p-AKT (Mann-Whitney U test) (Fig. 3c) and p-mTOR (Mann-Whitney U test) (Fig. 3d) proteins in the ΔPIK3R1-BT-474 cell line, compared to the equivalent WT line.
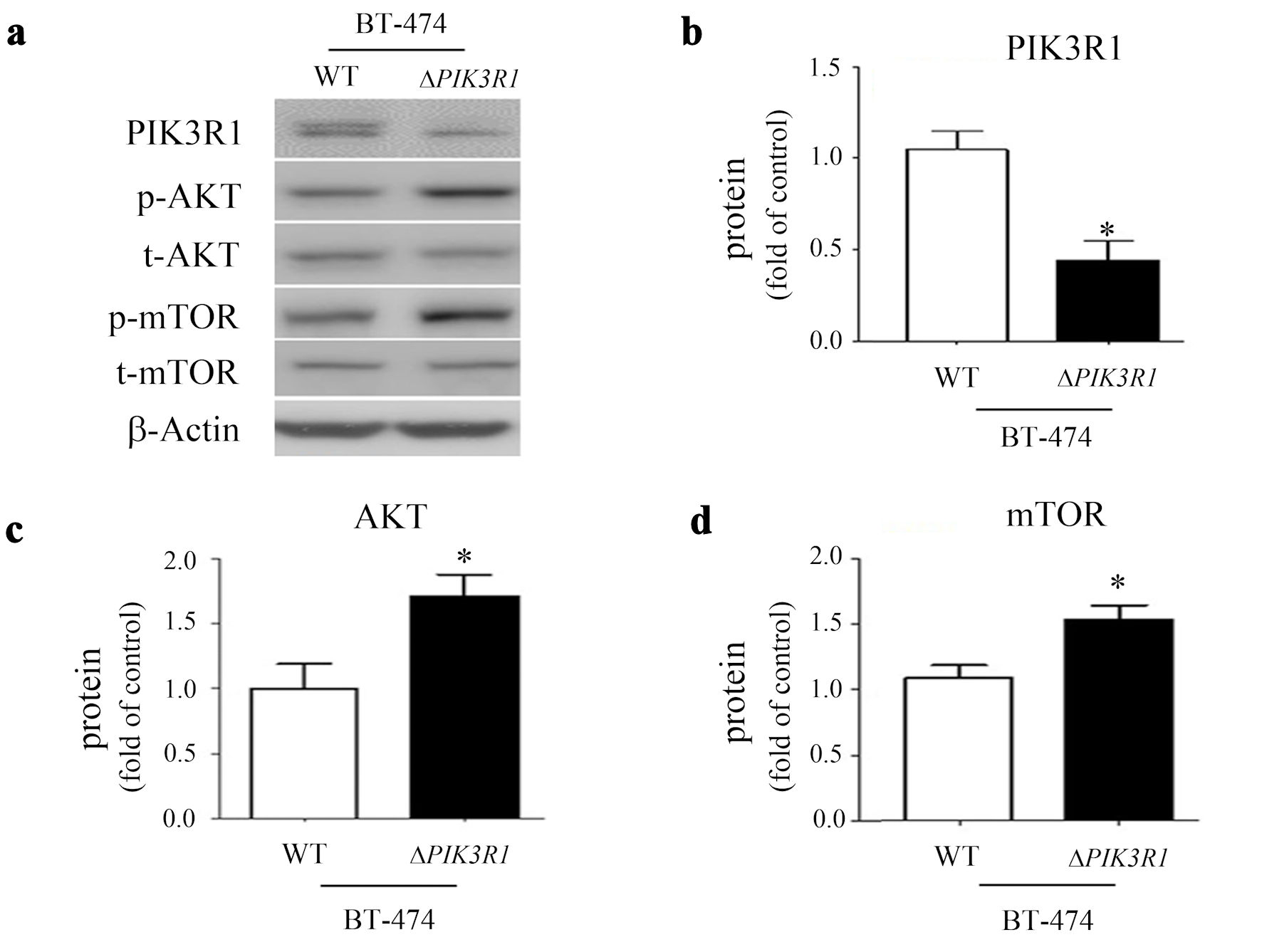 Click for large image | Figure 3. The role of PIK3R1 in cell growth signaling. Cell lysates obtained from BT-474 (both WT and ΔPIK3R1 cell lines) were subjected to Western blot analysis and probed with various primary antibodies, namely PIK3R1/p85, p-AKT, t-AKT, p-mTOR, mTOR and beta-actin (a). The protein expressions of PIK3R1/p85 (b), AKT (c) and mTOR (d) were quantified as a relative ratio (t-protein/p-protein) and presented as fold of control (ΔPIK3R1/WT). Asterisks indicate a P value < 0.05 by Mann-Whitney U test. WT: wild type. |
The expression of PIK3R1 in human normal breast and tumor tissue
Next, we investigated the PIK3R1 protein expression in human breast tissue, including normal breast and tumor tissue. After human breast tissue samples were stained with anti-PIK3R1 antibody, the positivity of PIK3R1 expression was semi-quantified (Fig. 4). The results demonstrated that normal breast tissue had a high level of PIK3R1 expression (3+), while luminal type breast cancer tumor samples showed a variety of different level of expression of PIK3R1 (ranging from 0+ to 3+) (Supplementary Material 3, wjon.elmerpub.com).
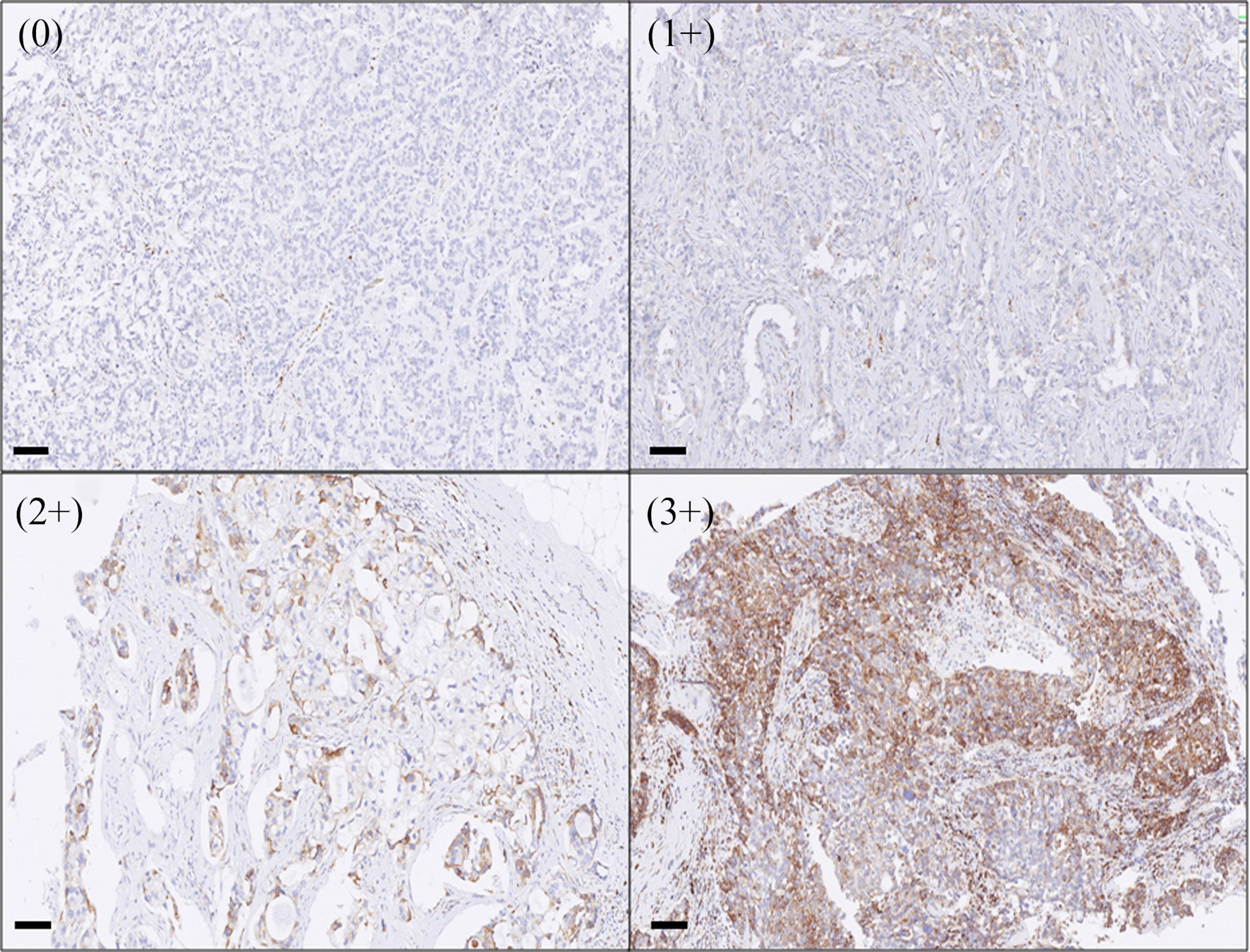 Click for large image | Figure 4. The expression of PIK3R1 in human breast cancer tumor tissue. When human breast tissues were stained with PIK3R1 antibody as described in the Methods, the positivity of the protein expression was semi-quantified and expressed as 0 (< 10%), 1+ (11-25%), 2+ (26-50%), 3+ (> 50% of cells examined). Bars = 100 µm. |
Correlation between PIK3R1 expression level and patient clinical outcomes
In order to elucidate the role of PIK3R1 in patients’ outcome, we performed IHC staining on surgically removed tumor tissues (breast cancer tissue array) with anti-PIK3R1 antibody and correlated the results with patient DFS/OS. Using Kaplan-Meier analysis to estimate patients’ outcomes, our results showed that patients with high expression of PIK3R1 protein (≥ 2+) had better outcomes, such as DFS (P = 0.038) and OS (P =0.034), compared to those with low expression of PIK3R1 protein (< 2+) in the early stages (stage I/II, n = 339) of breast cancer (Fig. 5a, b). In contrast, this correlation was not present across all stages of breast cancer (n = 440) in DFS (P = 0.182) (Fig. 5c) and OS (P = 0.142) (Fig. 5d).
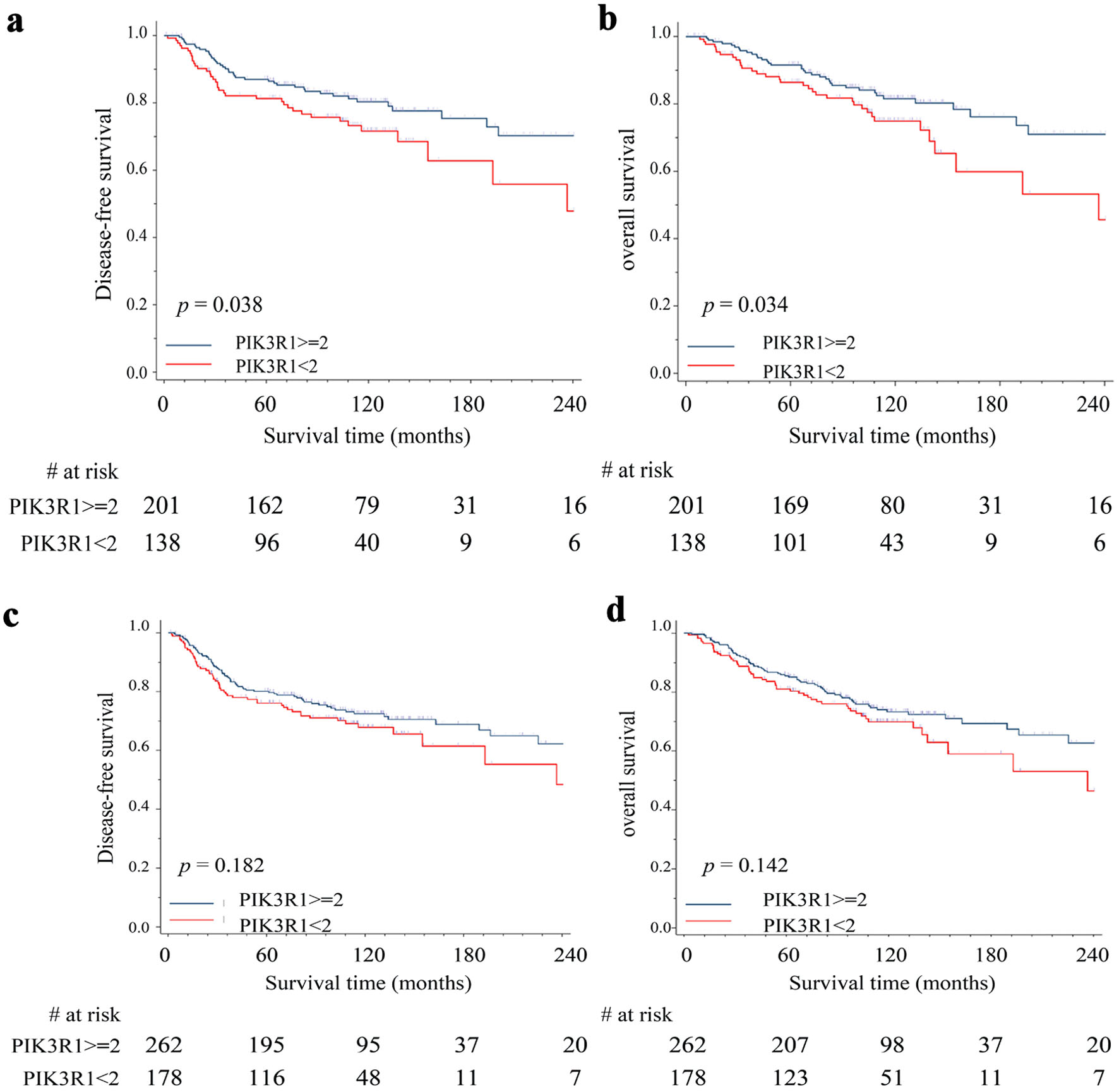 Click for large image | Figure 5. Correlation between PIK3R1 expression level and patient clinical outcomes. We performed immunohistochemical staining on surgically removed tumor tissues (breast cancer tissue array) with anti-PIK3R1 antibody and correlated the results with patient DFS/OS. Kaplan-Meier analysis was used to estimate patients’ outcome including DFS (a) and OS (b) in stage I/II patients (n = 339) and in all stage patients DFS (n = 440) (c) and OS (d). High expression of PIK3R1 protein indicates immunohistochemical staining score ≥ 2+; low expression of PIK3R1 protein indicates immunohistochemical staining score < 2+. A P value < 0.05 indicates statistical significance. DFS: disease-free survival; OS: overall survival. |
Correlation between PIK3R1 expression level and patient outcome by different breast cancer subtypes
Since breast cancer is a very heterogenous disease and has diverse pathological and biological patterns, we further analyzed the correlation between PIK3R1 expression and patient DFS/OS in early stage (n = 339) of different breast cancer subtypes. The results showed that there was no significant difference between high expression (≥ 2+) and low expression (< 2+) of PIK3R1 protein in patient DFS (P = 0.467) (Fig. 6a) and OS (P = 0.437) (Fig. 6b) in luminal type (n = 43) breast cancer. It is of note that patients with high expression of PIK3R1 protein (≥ 2+) had significantly better OS (P = 0.038), but not DFS (P = 0.252), compared to those with low expression of PIK3R1 protein (< 2+) in HER2-enriched (n = 162) breast cancer (Fig. 6c, d). In contrast, there was no statistical difference between high expression (> 2+) and low expression (< 2+) of PIK3R1 protein in triple-negative (n = 134) breast cancer, both in DFS (P = 0.388) (Fig. 6e) and OS (P = 0.697) (Fig. 6f).
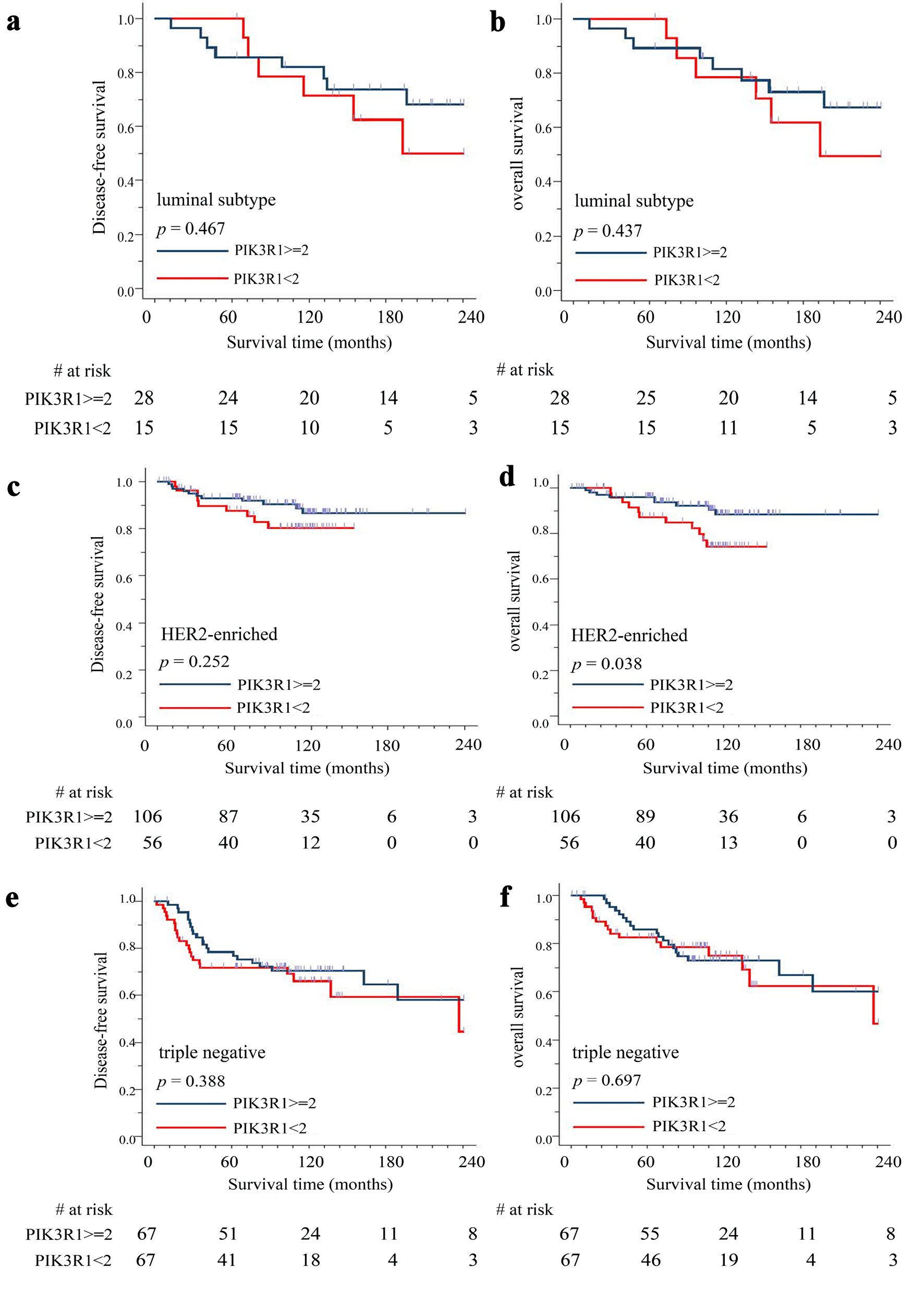 Click for large image | Figure 6. Correlation between PIK3R1 expression level and patient outcomes by different breast cancer subtypes. We performed immunohistochemical staining on surgically removed tumor tissues (breast cancer tissue array) with anti-PIK3R1 antibody and correlated the results with patient DFS/OS. Kaplan-Meier analysis was used to estimate stage I/II patients’ outcome (n = 339) including disease-free survival (DFS) and overall survival (OS) in luminal subtype DFS (n = 43) (a) and OS (b), HER2-enriched subtype DFS (n = 162) (c) and OS (d), and triple-negative subtype DFS (n = 134) (e) and OS (f). High expression of PIK3R1 protein indicates immunohistochemical staining score ≥ 2+; low expression of PIK3R1 protein indicates immunohistochemical staining score < 2+. A P value < 0.05 indicates statistical significance. |
| Discussion | ▴Top |
It is generally accepted that human cancer is considered a genetic disease and that mutations that affect oncogenes and/or tumor suppressor genes are the initiators of the oncogenesis process. Since Samuels et al discovered the PIK3CA mutations in breast cancer, accumulating evidence has suggested that changes to the PIK3CA gene sequence are one of the most common actionable mutations [14-16], and identification of such mutations provides a molecular explanation in treatment resistance of human breast cancer. In our cohort database study, the prevalence of PIK3CA mutations in all samples and in HER2 (+) samples is 38% and 43%, respectively [23]. It is of note that, when patients are treated with cyclin-dependent kinase (CDK)4/6 inhibitors, the median time to treatment failure is 12 months for the group that is positive for a PIK3CA mutation, compared to 16 months for the group that is negative for a PIK3CA mutation, with the 95% confidence interval (CI) values being 7 - 21 months and 11 - 23 months, respectively [24]. Nonetheless, there is consensus that mutation in the PIK3CA gene still cannot explain treatment resistance in those patients with a PIK3CA mutation.
The PIK3R (phosphoinositide-3-kinase regulatory subunit) includes PIK3R1, PIK3R2, and PIK3R3. PIK3R1 is well recognized as a tumor suppressor, while PIK3R2 is able to act as a tumor driver during the tumorigenesis process [31, 32]. The PIK3R1 gene mutations are highly associated with a number of human diseases, including SHORT syndrome [18] and PI3Kδ syndrome 2 [19]. Besides, PIK3R1 gene mutations are known to result in abnormal activity of the PI3K-Akt-mTOR pathway. Somatic PIK3R1 variation is known to be associated with vascular malformations and overgrowth [33], while miR-100-3p is recognized to inhibit the adipogenic differentiation of human mesenchymal stem cells (hMSCs) by targeting PIK3R1 via the PI3K/AKT signaling pathway [34].
There is evidence that mutations in PIK3R1 result in cancer cell proliferation, and they are now identified as actionable mutations in ovarian carcinoma [20] and breast carcinoma [26]. Furthermore, in a previous study, a higher sample frequency of PIK3R1 mutations was observed in metastatic prostate cancers compared to primary prostate cancers [21]. It should be noted that when our patient cohort database is examined, we found that there was a higher frequency of PIK3R1 mutation in the pathological complete response (PCR) group compared to non-pathological complete response (non-PCR) group for those patients who were receiving neo-adjuvant systemic treatment (Supplementary Material 4, wjon.elmerpub.com). Using The Cancer Genome Atlas (TCGA) database, we stratified the breast cancer into luminal, HER2-enriched, and triple-negative subtypes. The results showed that there is no correlation between PIK3R1 gene mutations and clinical outcomes, including DFS and OS, in luminal, HER2-enriched, and triple-negative breast cancer subtypes (Supplementary Material 5, wjon.elmerpub.com).
Being a heterogeneous disease, breast carcinoma contains many subtypes. It is not surprising to observe that there is diverse PIK3R1 expression in different breast carcinoma cell lines (Supplementary Material 2, wjon.elmerpub.com). A previous investigation has demonstrated that berberine (BBR) inhibits cell growth of both MDA-MB-231 and MDA-MB-468 cells via different mechanisms, which indicates that the cellular responses to BBR treatment depend on molecular subtypes of the breast carcinoma [35]. In addition, recent evidence indicates that endoplasmic reticulum is able to activate many receptor tyrosine kinases including HER2/neu (HER2) and epidermal growth factor receptor (EGFR) [36]. These findings explain a possible mechanism for tamoxifen resistance in ER-positive/HER2-positive breast cancers [37]. It has been documented that an examination of the cross talk between endoplasmic reticulum and HER2 signaling might help identify new therapeutic strategies, such as the use of aromatase inhibitors, in dual blockade (trastuzumab/pertuzumab) treatment, as well as the use of CDK4/6 inhibitors. These strategies may be able to be used to treat a range of different breast cancer subtypes [38-40].
Using the Kaplan Meier plotter database, we found that there is a better OS in high PIK3R1 mRNA expression level (212240_s_at) compared to those with low PIK3R1 mRNA expression tumors in luminal A (ER+/HER2 low) subtype (P = 0.0018). There was a trend of positive correlation between PIK3R1 mRNA expression (212240_s_at) in patient OS in luminal B (ER+/HER2 high, P = 0.27) and HER2-enriched (P = 0.49) subtypes. In contrast, no correlation between PIK3R1 mRNA expression (212240_s_at) in patient OS was noticed in triple-negative subtype (P = 0.84) (Supplementary Material 6, wjon.elmerpub.com). Our in vitro data showed that there was a significant decrease in the G0G1-fraction and increased S-fraction, but not the S + G2M-fraction and cell growth signaling such as AKT/mTOR expression in ΔPIK3R1-MCF-7 cell line compared to WT cell line (Supplementary Material 7, wjon.elmerpub.com). We attribute this phenomenon to relative low knock down efficiency in MCF-7 line compared to BT-474 line. Finally, our results show that expression of the PIK3R1 protein is positively correlated with patient outcome for stage I and stage II breast cancers, suggesting that PIK3R1 plays an important role in tumor behavior such as cell proliferation; while factors involved in tumor metastasis such as epithelial mesenchymal transition, increased angiogenesis, and changes of microenvironment etc., contribute the tumor progression in late stage of breast cancer.
Conclusions
We conclude that downregulated PIK3R1 in BT-474 cells resulted in an increased cell growth and upregulated AKT-mTOR signaling. The high-expressed PIK3R1 protein in tumors correlates positively with patients’ outcome in stage I and II breast cancer.
| Supplementary Material | ▴Top |
Suppl 1. Validation of knock down efficiency of PIK3R1 gene on BT-474 breast cancer cell line.
Suppl 2. Expression of PIK3R1 in different human breast cancer cell lines.
Suppl 3. Expression of PIK3R1 in normal breast tissue and human breast cancers.
Suppl 4. Correlation between the presence of a PIK3R1 gene mutation and pathological complete response in human breast cancer patients.
Suppl 5. Correlation between PIK3R1 gene mutations and clinical outcomes in breast cancer subtypes.
Suppl 6. Correlation between PIK3R1 gene expression and clinical outcomes.
Suppl 7. The role of PIK3R1 on cell growth of the MCF-7 (ER+/HER2 low) cell line.
Acknowledgments
The authors wish to thank Mr. Morris Chang for his kind help during this study.
Financial Disclosure
This work was supported by grants from Taipei Veterans General Hospital (V112C-214; V113C-190; V114C-221) and Melissa Lee Cancer Foundation (MLCF_V112_A11204, V113_A11304).
Conflict of Interest
The authors have no relevant conflict of interest to disclose.
Informed Consent
Since the tumor tissues were de-linked from patients’ privacy, the consent for publication is not applicable.
Author Contributions
Chiu, JH and Tsai YF designed the study. Tsai YF and His CN drafted the manuscript. Hsu CY reviewed the pathological slides. Huang CC, Lai JI, Liu CY, Chao TC, Lin YS, Tsai YF, Feng CJ, and Tseng LM collected the subject’s data. Chiu JH and Tseng LM supervised the research. All authors reviewed and permitted the manuscript. Tsai YF and Chiu JH took full responsibility of execution and finalized the submitted manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article and its supplementary materials.
| References | ▴Top |
- Group AW, Crocetti E, Buzzoni C. [New incidence and mortality data. 2003-2005]. Epidemiol Prev. 2009;33(1-2):64.
pubmed - Ministry of Health and Welfare ROCT. Table 8 Cancer Causes of Death for Females. 2020.
- Viale G. The current state of breast cancer classification. Ann Oncol. 2012;23(Suppl 10):x207-x210.
doi pubmed - Guiu S, Michiels S, Andre F, Cortes J, Denkert C, Di Leo A, Hennessy BT, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol. 2012;23(12):2997-3006.
doi pubmed - Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492-2502.
doi pubmed - Sonnenblick A, Fumagalli D, Sotiriou C, Piccart M. Is the differentiation into molecular subtypes of breast cancer important for staging, local and systemic therapy, and follow up? Cancer Treat Rev. 2014;40(9):1089-1095.
doi pubmed - Sihto H, Lundin J, Lundin M, Lehtimaki T, Ristimaki A, Holli K, Sailas L, et al. Breast cancer biological subtypes and protein expression predict for the preferential distant metastasis sites: a nationwide cohort study. Breast Cancer Res. 2011;13(5):R87.
doi pubmed - Chiu JH, Wen CS, Wang JY, Hsu CY, Tsai YF, Hung SC, Tseng LM, et al. Role of estrogen receptors and Src signaling in mechanisms of bone metastasis by estrogen receptor positive breast cancers. J Transl Med. 2017;15(1):97.
doi pubmed - AlDubayan SH, Conway JR, Camp SY, Witkowski L, Kofman E, Reardon B, Han S, et al. Detection of pathogenic variants with germline genetic testing using deep learning vs standard methods in patients with prostate cancer and melanoma. JAMA. 2020;324(19):1957-1969.
doi pubmed - Mack PC, Banks KC, Espenschied CR, Burich RA, Zill OA, Lee CE, Riess JW, et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: Analysis of over 8000 cases. Cancer. 2020;126(14):3219-3228.
doi pubmed - Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017;168(4):584-599.
doi pubmed - Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274-1284.
doi pubmed - Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, Lisyanskaya A, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495-2505.
doi pubmed - Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554.
doi pubmed - Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, et al. PIK3CA mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle. 2009;8(9):1352-1358.
doi pubmed - Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21-41.
doi pubmed - Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125(4):733-747.
doi pubmed - Barcena C, Quesada V, De Sandre-Giovannoli A, Puente DA, Fernandez-Toral J, Sigaudy S, Baban A, et al. Exome sequencing identifies a novel mutation in PIK3R1 as the cause of SHORT syndrome. BMC Med Genet. 2014;15:51.
doi pubmed - Nguyen T, Lau A, Bier J, Cooke KC, Lenthall H, Ruiz-Diaz S, Avery DT, et al. Human PIK3R1 mutations disrupt lymphocyte differentiation to cause activated PI3Kdelta syndrome 2. J Exp Med. 2023;220(6):e20221020.
doi pubmed - D'Ambrosio C, Erriquez J, Arigoni M, Capellero S, Mittica G, Ghisoni E, Borella F, et al. PIK3R1(W624R) is an actionable mutation in high grade serous ovarian carcinoma. Cells. 2020;9(2):442.
doi pubmed - Chakraborty G, Nandakumar S, Hirani R, Nguyen B, Stopsack KH, Kreitzer C, Rajanala SH, et al. The impact of PIK3R1 mutations and insulin-PI3K-glycolytic pathway regulation in prostate cancer. Clin Cancer Res. 2022;28(16):3603-3617.
doi pubmed - Chen L, Yang L, Yao L, Kuang XY, Zuo WJ, Li S, Qiao F, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun. 2018;9(1):1357.
doi pubmed - Huang CC, Tsai YF, Liu CY, Chao TC, Lien PJ, Lin YS, Feng CJ, et al. Comprehensive molecular profiling of Taiwanese breast cancers revealed potential therapeutic targets: prevalence of actionable mutations among 380 targeted sequencing analyses. BMC Cancer. 2021;21(1):199.
doi pubmed - Chao TC, Tsai YF, Liu CY, Lien PJ, Lin YS, Feng CJ, Chen YJ, et al. Prevalence of PIK3CA mutations in Taiwanese patients with breast cancer: a retrospective next-generation sequencing database analysis. Front Oncol. 2023;13:1192946.
doi pubmed - Griffith OL, Spies NC, Anurag M, Griffith M, Luo J, Tu D, Yeo B, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun. 2018;9(1):3476.
doi pubmed - Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70.
doi pubmed - Craig DW, O'Shaughnessy JA, Kiefer JA, Aldrich J, Sinari S, Moses TM, Wong S, et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther. 2013;12(1):104-116.
doi pubmed - Chen JL, Wang JY, Tsai YF, Lin YH, Tseng LM, Chang WC, King KL, et al. In vivo and in vitro demonstration of herb-drug interference in human breast cancer cells treated with tamoxifen and trastuzumab. Menopause. 2013;20(6):646-654.
doi pubmed - Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-254.
doi pubmed - Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581-585.
doi pubmed - Liu Y, Wang D, Li Z, Li X, Jin M, Jia N, Cui X, et al. Pan-cancer analysis on the role of PIK3R1 and PIK3R2 in human tumors. Sci Rep. 2022;12(1):5924.
doi pubmed - Vallejo-Diaz J, Chagoyen M, Olazabal-Moran M, Gonzalez-Garcia A, Carrera AC. The Opposing Roles of PIK3R1/p85alpha and PIK3R2/p85beta in Cancer. Trends Cancer. 2019;5(4):233-244.
doi pubmed - Cottrell CE, Bender NR, Zimmermann MT, Heusel JW, Corliss M, Evenson MJ, Magrini V, et al. Somatic PIK3R1 variation as a cause of vascular malformations and overgrowth. Genet Med. 2021;23(10):1882-1888.
doi pubmed - Wang T, Zhong D, Qin Z, He S, Gong Y, Li W, Li X. miR-100-3p inhibits the adipogenic differentiation of hMSCs by targeting PIK3R1 via the PI3K/AKT signaling pathway. Aging (Albany NY). 2020;12(24):25090-25100.
doi pubmed - Lin YS, Chiu YC, Tsai YH, Tsai YF, Wang JY, Tseng LM, Chiu JH. Different mechanisms involved in the berberine-induced antiproliferation effects in triple-negative breast cancer cell lines. J Cell Biochem. 2019;120(8):13531-13544.
doi pubmed - Razandi M, Pedram A, Park ST, Levin ER. Proximal events in signaling by plasma membrane estrogen receptors. J Biol Chem. 2003;278(4):2701-2712.
doi pubmed - Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, Schiff R. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926-935.
doi pubmed - Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29(2):217-233.
doi pubmed - Giuliano M, Hu H, Wang YC, Fu X, Nardone A, Herrera S, Mao S, et al. Upregulation of ER signaling as an adaptive mechanism of cell survival in HER2-positive breast tumors treated with anti-HER2 therapy. Clin Cancer Res. 2015;21(17):3995-4003.
doi pubmed - Vigano L, Locatelli A, Ulisse A, Galbardi B, Dugo M, Tosi D, Tacchetti C, et al. Modulation of the Estrogen/erbB2 receptors cross-talk by CDK4/6 inhibition triggers sustained senescence in estrogen receptor- and ErbB2-positive breast cancer. Clin Cancer Res. 2022;28(10):2167-2179.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.