| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 000, Number 000, September 2025, pages 000-000
Invasive Lobular Carcinoma Has Higher Immune Response Than Invasive Ductal Carcinoma in Estrogen Receptor-Positive/Human Epidermal Growth Factor Receptor 2-Negative Breast Cancers
Gabrielle Yeea, b, g , Rongrong Wua, c, g, Takashi Ishikawac, Kazuaki Takabea, b, c, d, e, f, h
aBreast Surgery, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA
bDepartment of Surgery, Jacobs School of Medicine and Biomedical Sciences, State University of New York, Buffalo, NY, USA
cDepartment of Breast Surgery and Oncology, Tokyo Medical University, Tokyo, Japan
dDepartment of Gastroenterological Surgery, Yokohama City University Graduate School of Medicine, Yokohama, Japan
eDepartment of Surgery, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
fDepartment of Breast Surgery, Fukushima Medical University School of Medicine, Fukushima, Japan
gThese authors contributed equally to this work.
hCorresponding Author: Kazuaki Takabe, Breast Surgery, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA
Manuscript submitted January 3, 2025, accepted August 14, 2025, published online September 13, 2025
Short title: Higher Immune Response in ILC vs. IDC in ER+/HER2- BC
doi: https://doi.org/10.14740/wjon2529
| Abstract | ▴Top |
Background: Invasive lobular carcinoma (ILC) and invasive ductal carcinoma (IDC) are two major pathological diagnoses of breast cancer, but few studies have described their differences within luminal (estrogen receptor (ER)-positive/human epidermal growth factor receptor 2 (HER2)-negative) subtypes at the molecular level.
Methods: Using The Cancer Genome Atlas (TCGA) (n = 584) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (n = 1,355) cohorts, we analyzed luminal ILC and IDC, excluding mixed type, in patients with stage I-III breast cancer.
Results: ILC was associated with Nottingham histological grade 2, larger tumor size and more stage III disease than IDC (all P < 0.01) but no difference in lymph node nor distant metastasis in both cohorts. There was no survival difference between ILC and IDC. ILC had less aggressive genomic features compared to IDC, and the cell proliferation score and Ki67 gene expression were significantly lower in ILC in TCGA (P < 0.001); however, these findings were not validated in METABRIC. Hallmark cell proliferation-related gene sets (E2F targets, G2M checkpoint, MYC targets V1, and MTORC1 signaling) were significantly less enriched in ILC in both cohorts (all normalized enrichment score (NES) > 1.4, false discovery rate (FDR) < 0.12). While ILC appeared to have a lower trend of pathological complete response (pCR) in the GSE20194 and GSE1140494 cohorts, ILC was infiltrated with significantly more CD4+ cells and dendritic cells and significantly less T helper type I (Th1) cells, regulatory T cells and M1 and M2 macrophages in both cohorts (all P < 0.05). Stromal cells, adipocytes and lymphatic endothelial cells were highly infiltrated in ILC, and cytolytic activity that represented the global anti-tumor immunity was significantly elevated in ILC in TCGA and subsequently validated in METABRIC.
Conclusions: ILC has higher immune response and immune cell infiltration than IDC in the luminal subtype.
Keywords: Breast cancer; Invasive lobular carcinoma; Invasive ductal carcinoma; Gene expression; Cell proliferation; Immune response
| Introduction | ▴Top |
The importance of tumor biology in both the staging and treatment of breast cancer patients has become paramount [1, 2]. Most breast cancers are pathologically classified as either invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC) or mixed type. It is clinically recognized that wide variations in outcome exist among patients with cancers with differing histologic features [3]. Approximately 10-15% of all breast cancers are characterized as ILC and are identified pathologically by small, round tumor cells surrounded by stroma growing in a discohesive single-file pattern [4, 5]. A retrospective analysis by Danzinger et al of 485 patients with primary ILC or IDC found that ILC were more likely to be tumor grade 2, estrogen receptor (ER)-positive, have lower proliferation rate and a larger size tumor compared to IDC [6].
Additionally, ILC and IDC have molecular differences. To paint a molecular portrait, several studies have analyzed the genomic, transcriptomic and proteomic pathways activated in ILC [7-9]. For example, Zhao et al found differing global transcription patterns between ILC and IDC, and Kurozumi et al found targetable ERBB2 mutations enriched in primary ILC [10, 11]. However, many of these previous studies compared all types of ILC and IDC, even though it is known that majority of ILC are the ER-positive subtype. Therefore, the previous publications may have observed the difference between ER-positive cancers and the other subtypes. Little is known about the molecular variability between ILC and IDC specifically among the ER-positive/human epidermal growth factor receptor 2 (HER2)-negative subtype.
To date, our group has been pursuing in silico translational research to study the clinical relevance of gene expression in cancer [12-17]. We have analyzed the biology of pathological findings to see if pathologically determined aspects of cancer reflect the underlying molecular features of tumors and if these features can be discovered within their genomic and transcriptomic portraits. For instance, we reported that breast cancers with lymphovascular invasion are associated with high proliferation, but not lymphangiogenesis nor immune response [18]. We have also shown that Nottingham histological grade 3 breast cancers have not only enhanced cell proliferation but also aggressive transcriptomic features with enhanced immunogenicity and elevated T-cell exhaustion markers [19]. Furthermore, we found that metaplastic breast cancer has increased intratumor heterogeneity, enriched angiogenesis and epithelial-mesenchymal transition (EMT) gene sets, as well as being associated with worse disease-specific survival [20]. Lastly, we have found that a high proportion of intratumoral adipocytes in breast cancer are associated with inflammation, metastatic pathways, cancer stemness and favorable tumor immune microenvironment; however, a low proportion of adipocytes were associated with aggressive cancer cell proliferation in ER-positive breast cancer [21].
To this end, we hypothesized that ER-positive/HER2-negative ILC and IDC are biologically distinct, in that ILC is associated with less cell proliferation and subsequent worse response to neoadjuvant chemotherapy compared with IDC. Our findings may help explain some clinical features of ILC and hint at novel therapeutic options.
| Materials and Methods | ▴Top |
Breast cancer patient cohorts
Two publicly available datasets were collected from online sources and analyzed in this study. As the testing cohort, we used the Pan Cancer Clinical Data Resource [22] and the cBio Cancer Genomic Portal [23] to obtain data regarding tumor gene expression and clinical data from The Cancer Genome Atlas (TCGA) cohort, which consists of over 11,000 patient samples, in the same manner as our group has previously reported [24, 25]. Among them, 584 patients who were ER-positive/HER2-negative by pathological determination were analyzed. Nottingham histological grade of primary breast tumors of TCGA was assessed from pathology reports as shown previously [19]. The validation cohort used that has gene expression and correlating clinical data was the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (n = 1,355), which was accessed using the cBio portal system, as our group has previously described [14, 26, 27]. The cohorts GSE20194 (n = 138) and GSE140494 (n = 51) were analyzed to assess the relationship with the response to neoadjuvant chemotherapy [28, 29]. These cohorts were downloaded from the GEO database via the R package GEOquary. GSE20194 is a cohort consisting of 230 patients with stage I-III breast cancer who received 6 months of neoadjuvant chemotherapy including paclitaxel, 5-fluorouracil, cyclophosphamide and doxorubicin followed by surgical resection of the cancer at the University at Texas M.D. Anderson Cancer Center [28, 30]. In this cohort, six patients with IDC and one patient with ILC achieved pathological complete response (pCR), and 111 patients with IDC and 20 patients with ILC had residual disease. GSE140494 is a cohort of 114 patients in Germany who were treated with 3-week cycles of docetaxel followed by 3-week cycles of 5-fluorouracil, epirubicin, and cyclophosphamide. In this cohort, seven patients with IDC and one patient with ILC achieved pCR, and 31 patients with IDC and 12 patients with ILC had residual disease [29]. All patient information in both cohorts was deidentified; therefore, Institutional Review Board approval at Roswell Park Comprehensive Cancer Center (Buffalo, NY, USA) was waived as publicly available deidentified databases were used. The study was conducted in compliance with the ethical standards of the responsible institution.
Gene set expression analysis
As performed in previous studies from our group, we performed Gene Set Enrichment Analysis (GSEA) [31], which is a computational method to investigate the biological status of each set of genes. GSEA analyzes the statistical significance between two groups in the expression of pre-defined gene collection (gene set) representing specific biological pathways. Gene sets used in this study were from the Hallmark and Pathway Interaction Database (PID) collection of Molecular Signatures Database (MSigDB) [32]. The analysis was performed with GSEA software and hallmark gene set collection for pathway analysis. We used a false discovery rate of 0.25, as previously recommended for GSEA.
Statistical analysis
The composition of infiltrating immune cells and stromal cells in the tumor microenvironment were estimated from gene expression data using xCell algorithm [33]. Statistical analyses and data plotting were performed using R software and Microsoft Excel. A P value threshold of 0.05 was used to declare statistical significance. To determine significance among different groups we used one-way analysis of variance (ANOVA) or Fisher’s exact, as we described in legends. For survival analysis, the Kaplan-Meier method with log-rank test was used.
| Results | ▴Top |
ILC has more Nottingham histologic grade 2, larger tumor size and more stage III disease compared to IDC within ER-positive/HER2-negative subtype
We analyzed the difference in American Joint Committee on Cancer (AJCC) cancer staging between IDC and ILC ER-positive/HER2-negative breast cancers in the TCGA and METABRIC cohorts. The Nottingham histological grade, which pathologically assesses cancer cell proliferation, was significantly lower (more grade 2 and less grade 3) in ILC consistently in both cohorts (both P < 0.01) (Fig. 1). ILC had more T3 tumors than IDC in the TCGA cohort, which is in agreement with the previous report that included all subtypes [6] (P < 0.001) (Fig. 1), and ILC was significantly associated with larger tumor size compared to IDC in METABRIC cohort, which does not provide T-category of AJCC staging (P < 0.02) (Fig. 1). We found no significant differences in lymph node or distant metastasis between IDC and ILC in either of the cohorts (Fig. 1). Despite no difference in N, nor M category, ILC had more stage III disease than IDC in both TCGA and METABRIC, but it was only significant in the former cohort (P < 0.001) (Fig. 1).
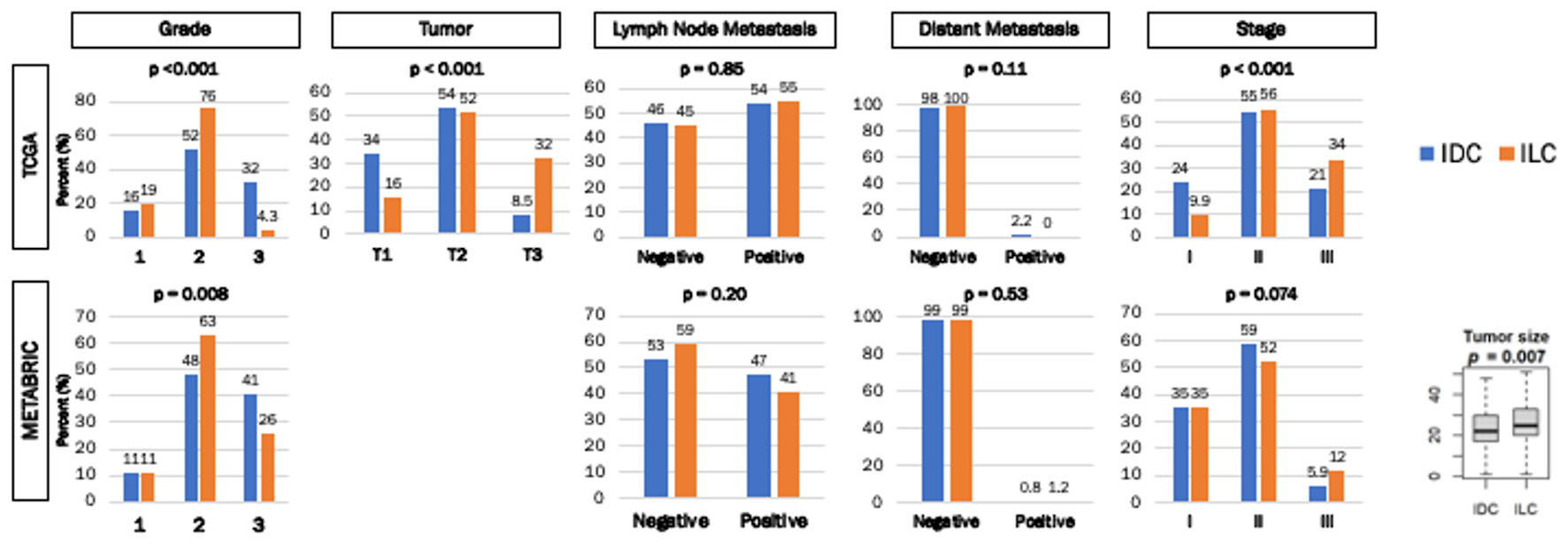 Click for large image | Figure 1. Clinical parameters of ER+/HER2- IDC and ILC breast cancers in TCGA and METABRIC cohorts. Bar graphs demonstrate the percentage of patients with histologic grades, tumor size, lymph node metastasis, distant metastasis, and stage. The Y-axis demonstrates the percentage of patients in either cohort. The X-axis represents grade (1, 2 and 3), tumor size (T1, T2, T3 and T4), patients with and without lymph node metastasis, patients with and without distant metastasis, and stage (I, II and III) in each cohort. Box plot demonstrates tumor size difference between IDC and ILC in the METABRIC cohort. Numbers above each bar represent the percentage of patients within each cohort. IDC were represented by the blue bars, and ILC were represented by the orange bars. Fisher’s exact test was used to calculate P values. |
There was no disease-free survival (DFS), disease-specific survival (DSS), or overall survival (OS) difference between IDC and ILC of ER-positive/HER2-negative subtype
Given the difference in grade, tumor size and AJCC stage between ILC and IDC, we assessed the differences in survival outcomes between the two in TCGA and METABRIC cohorts. We found that there was no significant difference between IDC and ILC in ER-positive/HER2-negative breast cancer patients in DFS, DSS, or OS within either TCGA or METABRIC (Fig. 2). Our finding agreed with a previous study, which implicates that our cohorts follow the same trend [34].
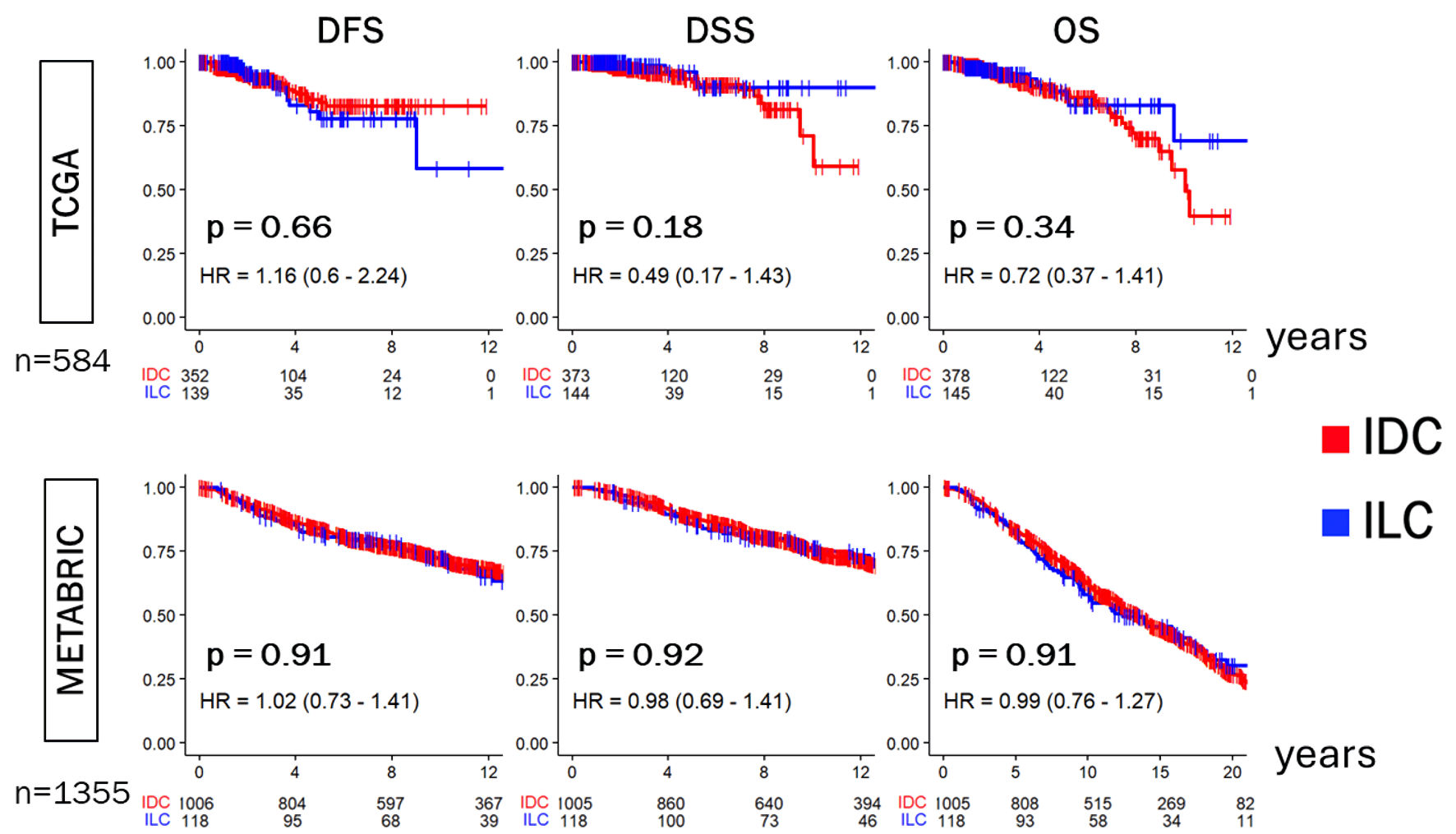 Click for large image | Figure 2. Disease-free survival (DFS), Disease-specific survival (DSS), and overall survival (OS) of ER-positive/HER2-negative IDC and ILC in the TCGA and METABRIC cohorts. Kaplan-Meier plots show DFS, DSS, and OS in the TCGA (n = 584) and METABRIC (n = 1,355) cohorts. Log-rank test was conducted to determine P values. IDC were indicated by the red lines, and ILC by the blue lines. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma; TCGA: The Cancer Genome Atlas; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; HR: hazard ratio. |
ILC was associated with significantly less intratumoral heterogeneity, homologous recombination deficiency (HRD), altered fractions and mutation rates
Because mutation load is known to correlate with cancer aggression [20, 35-38], we examined its association with IDC and ILC in ER-positive/HER2-negative subtype. Utilizing the scores pre-calculated by Thorsson et al [39, 40], we found that ILC was significantly associated with lower intratumoral heterogeneity, HRD, fraction altered, silent and non-silent mutation rates compared to that of IDC in TCGA (all P < 0.001) (Fig. 3a). Interestingly, there was no significant difference in either single nucleotide variant (SNV) or indel neoantigens between IDC and ILC (Fig. 3b).
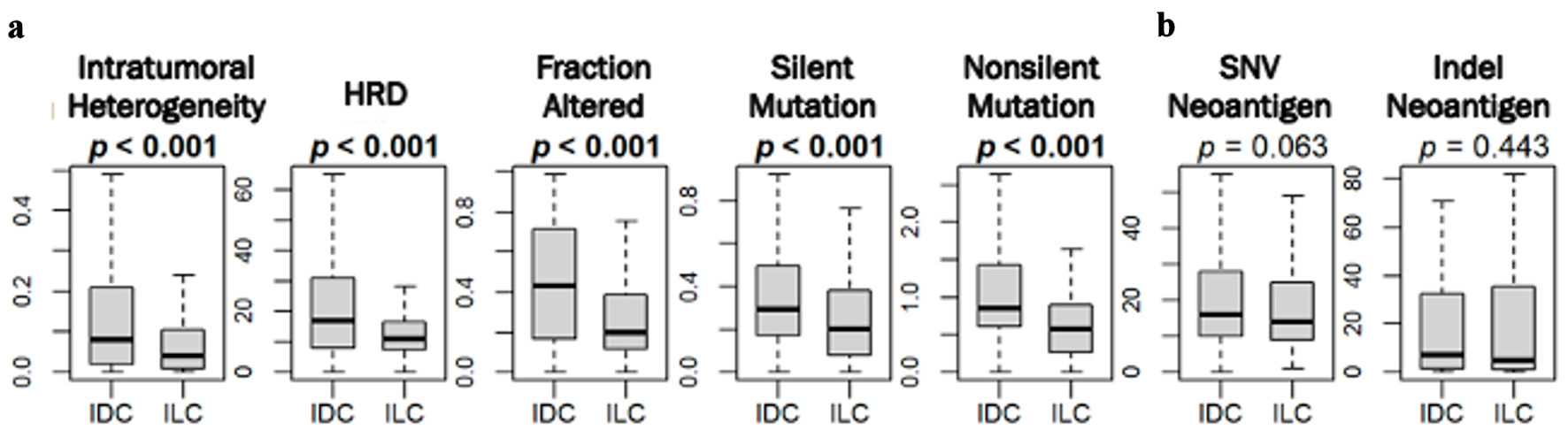 Click for large image | Figure 3. Mutation rates amongst ER-positive/HER2-negative IDC and ILC in TCGA. Box plots show (a) intratumoral heterogeneity, homologous recombination deficiency (HRD), fraction altered, silent and non-silent mutation rate, and (Bb single nucleotide variant (SNV) and indel neoantigens between IDC and ILC in the TCGA cohort. P values were determined using Mann Whitney U test. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma; TCGA: The Cancer Genome Atlas. |
ILC was less proliferative compared with IDC
Given the previous reports that ILC is less proliferative than IDC [6], it was of interest to compare the cell proliferation of patients with IDC and ILC ER-positive/HER2-negative breast cancer in TCGA and METABRIC cohorts. Gene expression of Ki67 (MKi67), one of the most used markers of cell proliferation in the clinical setting, and proliferation score were both significantly lower in ILC in TCGA (P < 0.001) (Fig. 4a,) but this was not validated in METABRIC.
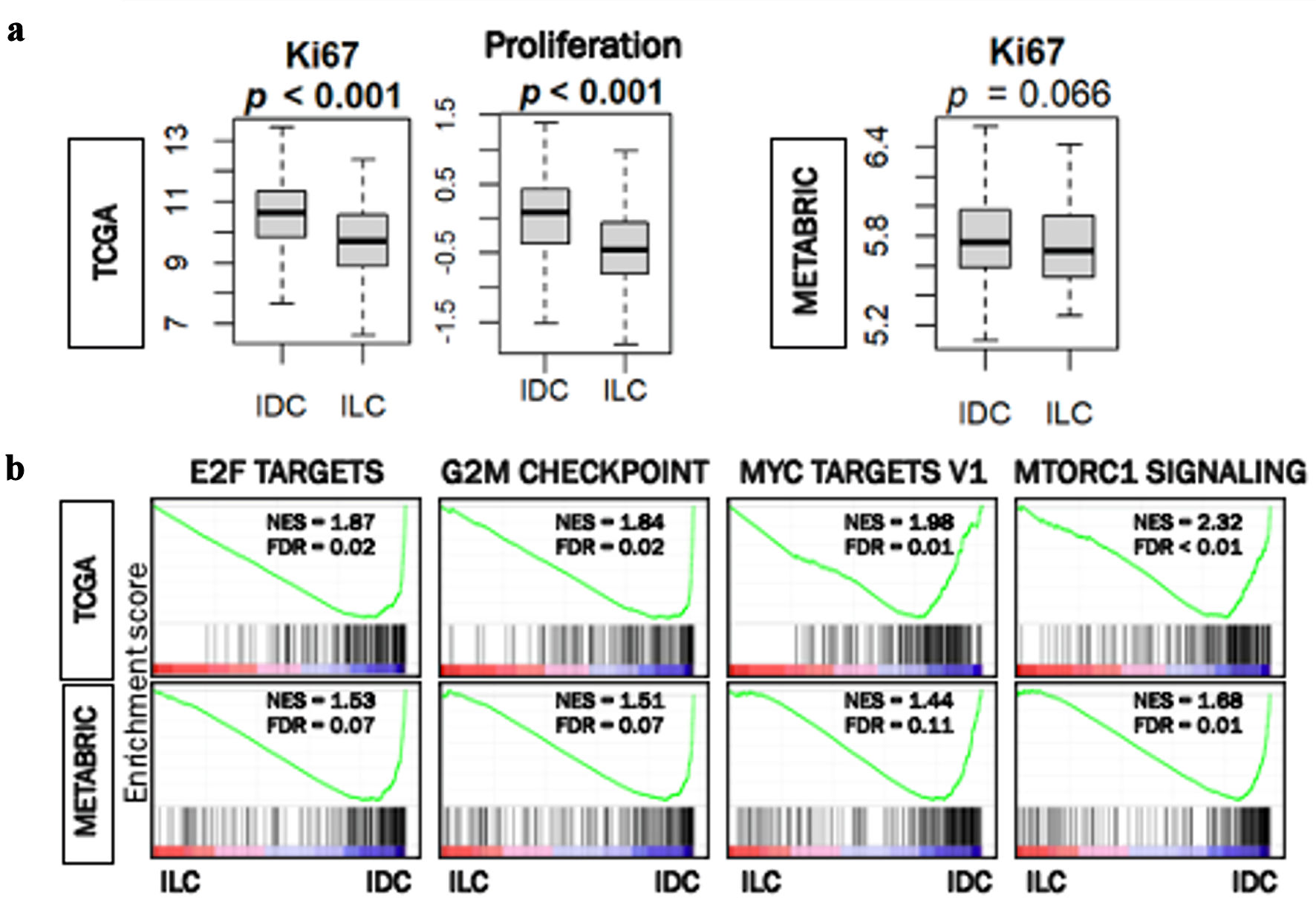 Click for large image | Figure 4. Cell proliferation rates in ER-positive/HER2-negative IDC and ILC in TCGA and METABRIC. (a) Box plots of MKi67 expression and proliferation score in IDC and ILC in TCGA cohort. Box plot of MiK67 expression in IDC and ILC in METABRIC cohort. Mann Whitney U test was used to calculate P values. (b) Gene set enrichment analysis (GSEA) of hallmark collection (E2F targets, G2M checkpoint, MYC targets V1, and MTORC1 signaling) and their relationship with IDC and ILC cells in the TCGA and METABRIC cohorts. Median cut-off was used to perform the analysis. NES and FDR were determined with the GSEA method. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma; TCGA: The Cancer Genome Atlas; METABRIC: Molecular Taxonomy of Breast Cancer International Consortium; NES: normalized enrichment score; FDR: false discovery rate. |
Likewise, we found that cell proliferation-related gene sets in the hallmark collection (E2F targets, G2M checkpoint, MYC targets V1, and mTORC1 signaling) were all significantly enriched in IDC consistently in both cohorts (Fig. 4b). These results uniformly suggest that ILC was associated with less cancer cell proliferation in TCGA and METABRIC cohorts.
No difference in response to neoadjuvant chemotherapy between IDC and ILC in GSEA20194 and GSE140494 cohorts
It is well known that proliferative cancer responds to cytotoxic chemotherapy better than slow growing tumors [41-43]. To this end, it was of interest to query the response of patients with ER-positive/HER2-negative IDC and ILC to neoadjuvant chemotherapy. Although IDC appeared to have a higher trend of pCR compared with ILC, it was not statistically significant neither in GSE20194 cohort (5.1% versus 4.8%, P > 0.99), nor in the GSE140494 cohort (18% and 7.7%, P = 0.66) (Fig. 5).
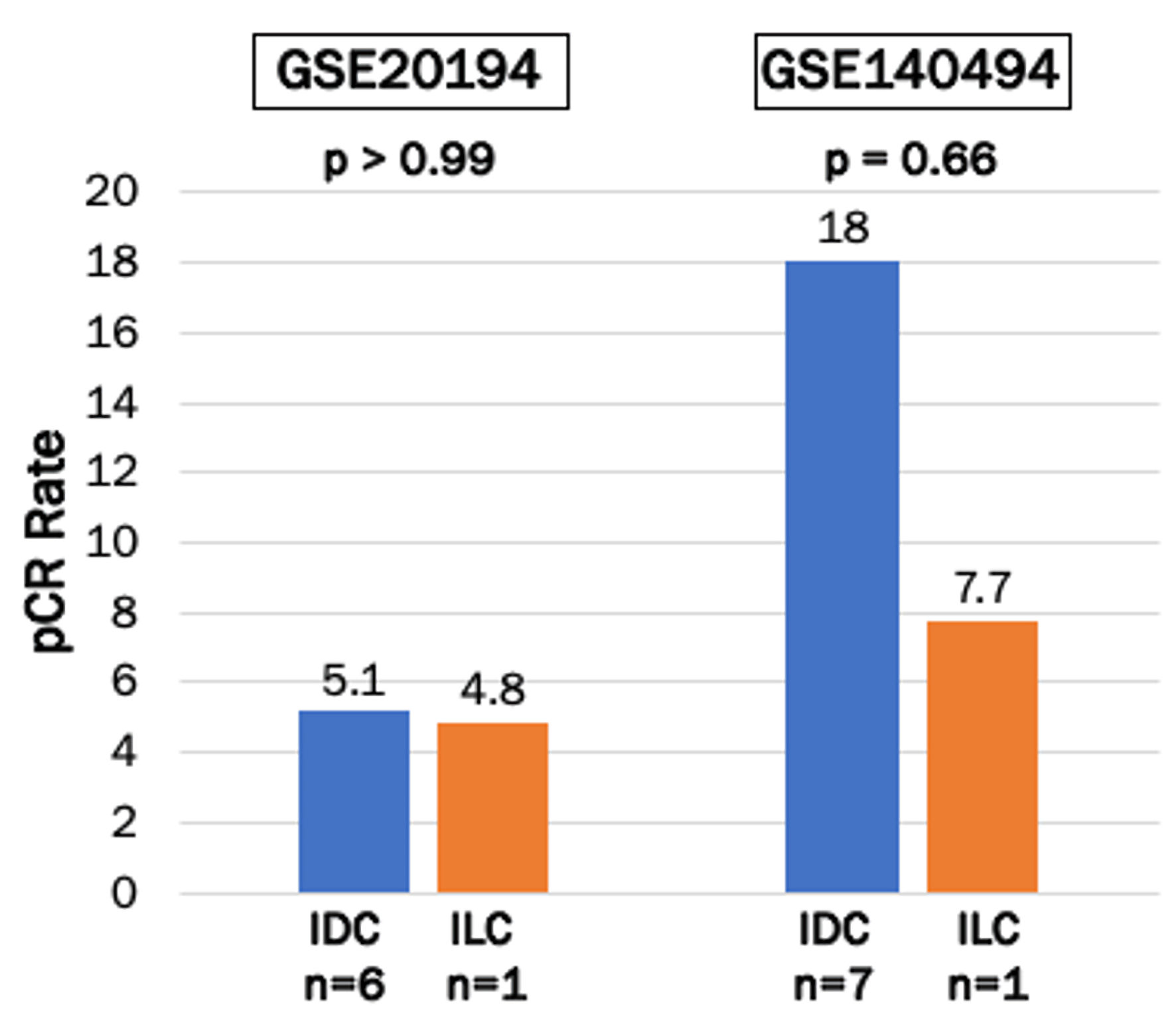 Click for large image | Figure 5. Pathological clinical response (pCR) to neoadjuvant chemotherapy in ER-positive/HER2-negative IDC and ILC in GSE20194 and GSE140494 cohorts. Bar plots show pCR rates after neoadjuvant chemotherapy in ER-positive/HER2-negative IDC and ILC breast cancers in GSE20194 (n = 138) and GSE140494 (n = 51) cohorts. IDC were represented by the blue bars, and ILC were represented by the orange bars. Fisher’s exact test was used to calculate P values. ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma. |
Immune and stromal cell infiltration and cytolytic activity (CYT) were significantly high in ILC
We then investigated the relationship of ER-positive/HER2-negative IDC and ILC and immune response in the tumor microenvironment. We found that ILC was significantly associated with higher expression of leukocyte fraction (P < 0.001) but not tumor infiltrating lymphocyte (TIL) regional fraction score in the TCGA cohort (Fig. 6a). Multiple anti-cancer immune cells, including naive CD4 T cells, terminally differentiated effector memory CD4+ T (Tem) cells, Th1 cells, and conventional dendritic cells (cDC) were significantly infiltrated in ILC (all P < 0.02 in both cohorts) (Fig. 6b). In contrast, pro-cancer immune cells, regulatory T cells (Treg), and both M1 and M2 macrophages were significantly less infiltrated in ILC (all P < 0.04 in both cohorts) (Fig. 6b).
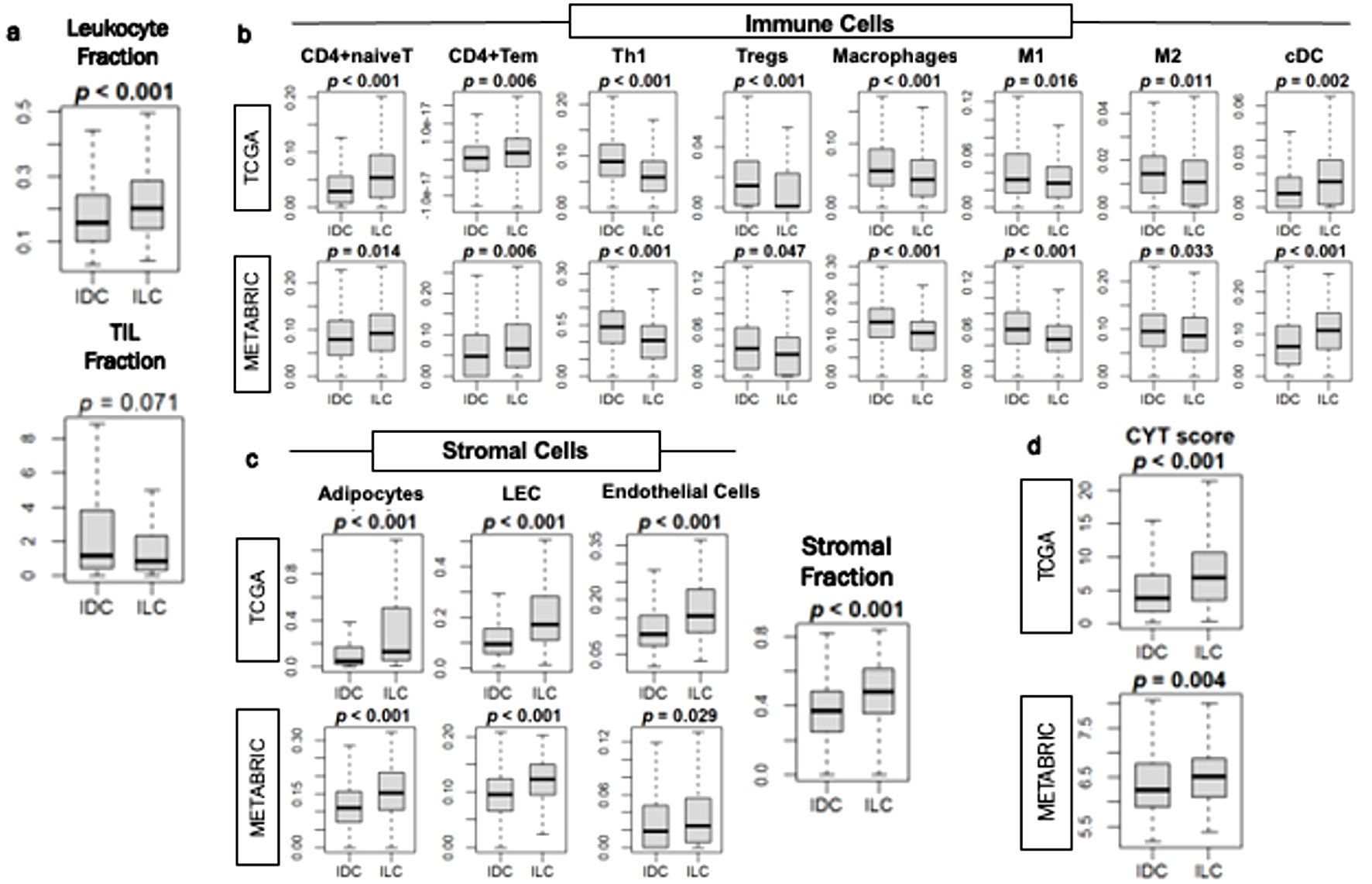 Click for large image | Figure 6. Immune response in ER-positive/HER2-negative IDC and ILC breast cancer tumor microenvironment. (a) Box plots of leukocyte fraction and tumor-infiltrating lymphocytes (TIL) regional fraction in the TCGA cohort. (b) Box plots of immune cells, including naive CD4 T cells (CD4+naiveT), effector memory CD4 T cells (CD4+Tem), T helper type 1 (Th1), regulatory T cells (Tregs), macrophages, M1 and M2 macrophages, and dendritic cells (cDC), in the TCGA and METABRIC cohorts. (c) Box plot of stromal cells, including adipocytes, lymphatic endothelial cells (LEC) and endothelial cells in the TCGA and METABRIC cohorts. Box plot of stromal fraction using the TCGA cohort. (d) Box plots of cytolytic activity (CYT) score in TCGA and METABRIC cohorts. Mann-Whitney U test was used to determine P values. IDC: invasive ductal carcinoma; ILC: invasive lobular carcinoma. |
Given the significant infiltration of immune cells and higher CYT in ILC, it was of interest to pursue a deeper analysis of specific immune pathways, such as expressions of immune checkpoint molecules and activation of immune-related signaling pathways. Although expressions of programmed cell death protein 1 (PD-1), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), lymphocyte-activation gene 3 (LAG3), indoleamine 2,3-dioxygenase 1 (IDO1), and T-cell immunoglobulin and ITIM domain (TIGIT) were significantly higher in ILC in TCGA, none of them were validated in METABRIC (Supplemental Material 1, wjon.elmerpub.com). Furthermore, we conducted GSEA between ILC and IDC. Among the hallmark collection gene sets, we found that the cell proliferation-related, glycolysis and unfolded protein response were the only gene sets enriched in IDC, and none were enriched in ILC, including immune-related sets such as allograft rejection, interferon (IFN)-alpha response, IFN-gamma response, interleukin (IL)2 signaling, IL6 signaling, and complement (data not shown).
Previously, we have shown that stromal cells such as adipocytes and lymphatic endothelial cells are highly infiltrated in a less proliferative tumor microenvironment [21, 44, 45]. Similarly, the infiltration of stromal cells such as adipocytes, lymphatic endothelial cells and endothelial cells, as well as the stromal fraction score, were all found to be significantly higher in ILC consistently in both cohorts (all P < 0.003) (Fig. 6c). CYT, which reflects overall anti-cancer immunity, was significantly higher in ILC compared to IDC consistently in both cohorts (P < 0.005) (Fig. 6d). These results suggest that ILC is less proliferative but has a more favorable tumor immune microenvironment compared to IDC.
| Discussion | ▴Top |
In this study, we used high-powered breast cancer patient cohorts TCGA and METABRIC to investigate the molecular differences between ER-positive/HER2-negative IDC and ILC breast cancers. We found that ILC was more often Nottingham histologic grade 2, larger in tumor size, and had more stage III and less stage I disease compared to IDC, although there was no difference in lymph node or distant metastasis. We demonstrated that there was no difference in DFS, DSS, or OS in both cohorts between IDC and ILC. Interestingly, ILC was associated with not only lower mutations rates, but also found to be less aggressive than IDC. ILC was also less proliferative compared with IDC measured by MKi67 expression, proliferation score and GSEA. There was no statistical difference between ILC and IDC in pCR rate after neoadjuvant chemotherapy in the GSE20194 and GSE140494 cohorts, either. We found that anti-cancer immune cell infiltrations and CYT were significantly higher in ILC. Given the fact that highly proliferative cancers, as well as cancers with anti-cancer immune cell infiltration respond to chemotherapy better, we cannot help but speculate that IDC and ILC divided these traits that resulted in no difference in response to neoadjuvant chemotherapy, followed by the survival outcomes.
First, we investigated whether the cohorts we used share similar clinical features with the previous studies. We found that there was a higher percentage of T3 tumors in ILC compared to IDC in TCGA, and the tumor size of ILC was found to be significantly larger than that of IDC in the METABRIC cohort both within ER-positive/HER2-negative subtype. This is in agreement with multiple previous studies that ILC are likely to be associated with larger tumor size [4, 6, 46, 47]. We found no significant difference in lymph node metastasis or distant metastases in TCGA or METABRIC, which is similar to the report by Danzinger et al [6]. We also found that ILC had more stage III disease than IDC in both TCGA and METABRIC, which is in agreement with previous studies that ILC is detected at a more advanced stage than IDC [48]. It is thought that ILC is found only after it grows to a large size in a locally advanced stage because it often does not form a discrete palpable mass and is more difficult to detect through physical examination [49-51]. Kim et al in their investigation on tumor size and its relation to immunotherapy response in murine models found that larger tumors are more immunosuppressive compared to small tumors [52]. Larger tumors may play a role in both the local and systemic effect, and this may suggest the relationship between immune cell infiltration and tumor burden.
Survival outcomes between IDC and ILC with positive hormone receptor have been shown to exhibit similar OS [4, 34]. Arpino et al studied the characteristics of ILC in 4,140 patients at Baylor and found similar 5-year DFS and OS with IDC [4]. In contrast, one single center study in China by Han et al demonstrated that the 5-year OS of ILC was worse than that of IDC after propensity score matching [53], and a large, retrospective analysis by Oesterreich et al found that patients with ER-positive ILC had statistically significant worse DFS and OS than ER-positive IDC [54]. Our study found no difference in DFS, DSS or OS between ER-positive/HER2-negative IDC and ILC in TCGA and METABRIC cohorts. A hint to explain this discrepancy may be in a report by Pramod et al [49], who noted that the incidence of ILC is different amongst varying races. When comparing incidence of ILC from the Surveillance, Epidemiology and End Results (SEER) database, they found that the 5-year OS and DSS were worse in Black patients compared with White women and women of other races [49]. While this was a much larger study, it did not compare ILC and IDC within the ER-positive/HER2-negative subtype. We know that TCGA sampled patients from the United States, and METABRIC from United Kingdom and Canada, and the analyses may warrant considering varying races in future studies.
Using multiple methods, we demonstrated that ER-positive/HER2-negative IDC was highly proliferative compared to that of ILC (Fig. 4). In both TCGA and METABRIC, ILC was associated with significantly lower Nottingham histological grade (less grade 3, both P < 0.01) (Fig. 1). MKi67, gene expression of one of the most commonly used markers of cell proliferation in clinics, and proliferation score were both significantly lower in ILC in TCGA (P < 0.001) (Fig. 4a). Additionally, the cell proliferation-related gene sets in the hallmark collection were all significantly enriched to IDC consistently in both cohorts (Fig. 4b). Mutation rates amongst ER-positive/HER2-negative ILC were found to be significantly less compared to IDC. Specifically, ILC was associated with significantly lower intratumoral heterogeneity, HRD, fraction altered, silent and non-silent mutation rate compared to that of IDC in TCGA (all P < 0.001) (Fig. 3a) . Interestingly, there was no significant difference in either SNV or indel neoantigens between IDC and ILC in our study (Fig. 3b). Together these results suggest that ILC in addition to being associated with lower mutation rates were also associated with less cancer cell proliferation.
In previous studies, we have shown that ER-positive/HER2-negative tumors with high E2F scores, i.e., highly proliferative tumors, achieved significantly higher pCR rate to neoadjuvant chemotherapy [41]. We have also shown that G2M score is associated with treatment response to systemic chemotherapy in ER-positive/HER2-negative cancer [42]. Therefore, given the strong relationship with cell proliferation, we anticipated that IDC would respond better to neoadjuvant chemotherapy. Although IDC was found to be more proliferative, there was no difference in pCR (Fig. 5). This is in agreement with the study by Delpech et al, who looked at 1,895 patients with ILC and IDC breast cancers and found that ILC was associated with a significantly lower pCR in univariate analysis but not significant after adjusting for tumor size and grade. There has been controversy about the benefit of chemotherapy in ILC. Some studies have shown significantly lower pCR in ILC compared to IDC after neoadjuvant chemotherapy [55-58]. However, these studies did not necessarily differentiate between ER-positive/HER2-negative subtype.
It is clear that the tumor microenvironment plays an important role in the progression of breast cancer. Although ER-positive/HER2-negative ILC was significantly associated with less mutations and cellular proliferation compared to that IDC, ILC was associated with significantly higher immune response in the tumor microenvironment (Fig. 6). ILC was significantly associated with higher leukocyte fraction (P < 0.001), and multiple anti-cancer immune cells were significantly infiltrative in ILC (all P < 0.02 in both cohorts) (Fig. 6b). Additionally, pro-cancer immune cells, regulatory T cells, helper type I T cells, and M1 and M2 macrophages were significantly less infiltrative in ILC (all P < 0.04 in both cohorts) (Fig. 6b). In a multicentric retrospective series of ER-positive/HER2-negative IDC and ILC tumors, Desmedt et al reported that TIL levels were statistically significantly lower in ILC compared with IDC. In ILC, high TIL levels were associated with young age, lymph node involvement, and high proliferative tumors [59]. Additionally, He et al also found that stromal scores of IDC were significantly lower than those of ILC [60]. The difference in immune cell infiltration findings between our study and these other studies may be due to the difference in the spatial locations of the samples studied. Our study analyzed the tissue within a tumor, as opposed to TIL which evaluated tissue at the edge of a tumor [61]. Furthermore, since we have observed higher immune cell infiltration in ILC consistently in two independent large breast cancer cohorts, we cannot help but speculate that our result may imply a potential role for immunotherapy for ILC, even in the ER-positive/HER2-negative subtype.
Although IDC was found to be more proliferative than ILC, perhaps we should not be misled by this association because ultimately there was no difference in pCR. Future biomarkers would be helpful to identify tumors that may or may not respond to neoadjuvant chemotherapy or endocrine therapy. Similar to LobSig, a molecular signature that captures the unique features of ILC, we hope that future prospective studies can identify molecular profiles that provide future prognostic information for patients with ILC [49, 62].
Our study has several limitations. First, this is a retrospective study using publicly available large patient cohorts TCGA and METABRIC containing clinical data and transcriptomes. These data have selection bias and are not necessarily reflective of the latest and most efficacious treatments within the time that they were collected. Additionally, our study does not include any in vitro or in vivo experiments. The results of our study are helpful for generating hypothesis and associations; however, future experiments will need to prove any potential underlying mechanisms.
In conclusion, in ER-positive/HER2-negative breast cancer, ILC has higher immune response and immune cell infiltration than IDC. ILC is also less proliferative and presents at a more advanced stage than IDC. There were no differences in achieving pCR after neoadjuvant chemotherapy nor survival differences between ILC and IDC.
| Supplementary Material | ▴Top |
Suppl 1. Gene expression levels of immune checkpoint molecules.
Acknowledgments
None to declare.
Financial Disclosure
This research was supported by the National Institutes of Health, USA, grant number R37CA248 -018, R01CA250412, R01CA251545, and R01EB029596; the US Department of Defense BCRP, grant number W81XWH-19-1-0674 and W81XWH-19-1-0111; and the National Institute of Health to Roswell Park, grant number P30CA016056 to K.T.
Conflict of Interest
None to declare.
Informed Consent
Not applicable.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Rongrong Wu. The first draft of the manuscript was written by Gabrielle Yee and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data Availability
The datasets analyzed in the current study were downloaded from cBioportal.
| References | ▴Top |
- Young J, Asaoka M, Ghasemi F, Chida K, Roy AM, Yan L, Hakamada K, et al. The American Joint Committee on Cancer (AJCC) breast cancer staging, eighth edition, is more reflective of cancer biology than the seventh edition. Ann Surg Oncol. 2025;32(5):3268-3277.
doi pubmed - Oshi M, Ghasemi F, Yamada A, Yan L, Zhang J, Abrams SI, Endo I, et al. Activated hippo pathway is associated with a worse response to trastuzumab and worse survival in HER2-positive breast cancer. Ann Surg Oncol. 2025.
doi pubmed - Tadros AB, Wen HY, Morrow M. Breast cancers of special histologic subtypes are biologically diverse. Ann Surg Oncol. 2018;25(11):3158-3164.
doi pubmed - Arpino G, Bardou VJ, Clark GM, Elledge RM. Infiltrating lobular carcinoma of the breast: tumor characteristics and clinical outcome. Breast Cancer Res. 2004;6(3):R149-156.
doi pubmed - Van Baelen K, Sawyer E, Van Cauwenberge J, Aftimos P, Covington MF, Maetens M, Zels G, et al. Clinical challenges and proposed solutions for patients with invasive lobular breast cancer. Ann Oncol. 2025.
doi pubmed - Danzinger S, Hielscher N, Izso M, Metzler J, Trinkl C, Pfeifer C, Tendl-Schulz K, et al. Invasive lobular carcinoma: clinicopathological features and subtypes. J Int Med Res. 2021;49(6):3000605211017039.
doi pubmed - Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506-519.
doi pubmed - Michaut M, Chin SF, Majewski I, Severson TM, Bismeijer T, de Koning L, Peeters JK, et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci Rep. 2016;6:18517.
doi pubmed - Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346-352.
doi pubmed - Kurozumi S, Alsaleem M, Monteiro CJ, Bhardwaj K, Joosten SEP, Fujii T, Shirabe K, et al. Targetable ERBB2 mutation status is an independent marker of adverse prognosis in estrogen receptor positive, ERBB2 non-amplified primary lobular breast carcinoma: a retrospective in silico analysis of public datasets. Breast Cancer Res. 2020;22(1):85.
doi pubmed - Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15(6):2523-2536.
doi pubmed - Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162(1):191-198.
doi pubmed - Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, et al. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel). 2020;12(10).
doi pubmed - Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, et al. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20(11).
doi pubmed - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, et al. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21(18).
doi pubmed - Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, et al. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC). Cancers (Basel). 2020;12(11).
doi pubmed - Yee G, Wu R, Oshi M, Endo I, Ishikawa T, Takabe K. Activity-regulated cytoskeleton-associated protein gene expression is associated with high infiltration of stromal cells and immune cells, but with less cancer cell proliferation and better overall survival in estrogen receptor-positive/human epidermal growth factor receptor 2-negative breast cancers. World J Oncol. 2025;16(1):16-29.
doi pubmed - Asaoka M, Patnaik SK, Zhang F, Ishikawa T, Takabe K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res Treat. 2020;181(2):309-322.
doi pubmed - Takahashi H, Oshi M, Asaoka M, Yan L, Endo I, Takabe K. Molecular biological features of nottingham histological grade 3 breast cancers. Ann Surg Oncol. 2020;27(11):4475-4485.
doi pubmed - Chouliaras K, Oshi M, Asaoka M, Tokumaru Y, Khoury T, Endo I, Ishikawa T, et al. Increased intratumor heterogeneity, angiogenesis and epithelial to mesenchymal transition pathways in metaplastic breast cancer. Am J Cancer Res. 2021;11(9):4408-4420.
pubmed - Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, et al. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21(16).
doi pubmed - Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400-416.e411.
doi pubmed - Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
doi pubmed - McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, Opyrchal M, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019;26(7):2191-2199.
doi pubmed - Katsuta E, Maawy AA, Yan L, Takabe K. High expression of bone morphogenetic protein (BMP) 6 and BMP7 are associated with higher immune cell infiltration and better survival in estrogen receptor-positive breast cancer. Oncol Rep. 2019;42(4):1413-1421.
doi pubmed - Yamada A, Nagahashi M, Aoyagi T, Huang WC, Lima S, Hait NC, Maiti A, et al. ABCC1-exported Sphingosine-1-phosphate, produced by Sphingosine Kinase 1, shortens survival of mice and patients with breast cancer. Mol Cancer Res. 2018;16(6):1059-1070.
doi pubmed - Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, et al. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20(17).
doi pubmed - Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28(8):827-838.
doi pubmed - Edlund K, Madjar K, Lebrecht A, Aktas B, Pilch H, Hoffmann G, Hofmann M, et al. Gene expression-based prediction of neoadjuvant chemotherapy response in early breast cancer: results of the prospective multicenter EXPRESSION trial. Clin Cancer Res. 2021;27(8):2148-2158.
doi pubmed - Popovici V, Chen W, Gallas BG, Hatzis C, Shi W, Samuelson FW, Nikolsky Y, et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5.
doi pubmed - Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550.
doi pubmed - Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425.
doi pubmed - Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220.
doi pubmed - Yang C, Lei C, Zhang Y, Zhang J, Ji F, Pan W, Zhang L, et al. Comparison of Overall Survival between invasive lobular breast carcinoma and invasive ductal breast carcinoma: a propensity score matching study based on SEER database. Front Oncol. 2020;10:590643.
doi pubmed - Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, et al. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10(1):1852.
doi pubmed - Oshi M, Gandhi S, Huyser MR, Tokumaru Y, Yan L, Yamada A, Matsuyama R, et al. MELK expression in breast cancer is associated with infiltration of immune cell and pathological compete response (pCR) after neoadjuvant chemotherapy. Am J Cancer Res. 2021;11(9):4421-4437.
pubmed - Oshi M, Kawaguchi T, Yan L, Peng X, Qi Q, Tian W, Schulze A, et al. Immune cytolytic activity is associated with reduced intra-tumoral genetic heterogeneity and with better clinical outcomes in triple negative breast cancer. Am J Cancer Res. 2021;11(7):3628-3644.
pubmed - Satyananda V, Oshi M, Endo I, Takabe K. High BRCA2 gene expression is associated with aggressive and highly proliferative breast cancer. Ann Surg Oncol. 2021;28(12):7356-7365.
doi pubmed - Sayaman RW, Saad M, Thorsson V, Hu D, Hendrickx W, Roelands J, Porta-Pardo E, et al. Germline genetic contribution to the immune landscape of cancer. Immunity. 2021;54(2):367-386.e368.
doi pubmed - Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830.e814.
doi pubmed - Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, et al. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9(7).
doi pubmed - Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, et al. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21(8).
doi pubmed - Oshi M, Gandhi S, Angarita FA, Kim TH, Tokumaru Y, Yan L, Matsuyama R, et al. A novel five-gene score to predict complete pathological response to neoadjuvant chemotherapy in ER-positive/HER2-negative breast cancer. Am J Cancer Res. 2021;11(7):3611-3627.
pubmed - Mukhopadhyay S, Tokumaru Y, Oshi M, Endo I, Yoshida K, Takabe K. Low adipocyte hepatocellular carcinoma is associated with aggressive cancer biology and with worse survival. Am J Cancer Res. 2022;12(8):4028-4039.
pubmed - Wu R, Sarkar J, Tokumaru Y, Takabe Y, Oshi M, Asaoka M, Yan L, et al. Intratumoral lymphatic endothelial cell infiltration reflecting lymphangiogenesis is counterbalanced by immune responses and better cancer biology in the breast cancer tumor microenvironment. Am J Cancer Res. 2022;12(2):504-520.
pubmed - Pestalozzi BC, Zahrieh D, Mallon E, Gusterson BA, Price KN, Gelber RD, Holmberg SB, et al. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26(18):3006-3014.
doi pubmed - Yeatman TJ, Cantor AB, Smith TJ, Smith SK, Reintgen DS, Miller MS, Ku NN, et al. Tumor biology of infiltrating lobular carcinoma. Implications for management. Ann Surg. 1995;222(4):549-559; discussion 559-561.
doi pubmed - Du T, Zhu L, Levine KM, Tasdemir N, Lee AV, Vignali DAA, Houten BV, et al. Invasive lobular and ductal breast carcinoma differ in immune response, protein translation efficiency and metabolism. Sci Rep. 2018;8(1):7205.
doi pubmed - Pramod N, Nigam A, Basree M, Mawalkar R, Mehra S, Shinde N, Tozbikian G, et al. Comprehensive review of molecular mechanisms and clinical features of invasive lobular cancer. Oncologist. 2021;26(6):e943-e953.
doi pubmed - Li CI, Uribe DJ, Daling JR. Clinical characteristics of different histologic types of breast cancer. Br J Cancer. 2005;93(9):1046-1052.
doi pubmed - DiCostanzo D, Rosen PP, Gareen I, Franklin S, Lesser M. Prognosis in infiltrating lobular carcinoma. An analysis of "classical" and variant tumors. Am J Surg Pathol. 1990;14(1):12-23.
doi pubmed - Kim SI, Cassella CR, Byrne KT. Tumor burden and immunotherapy: impact on immune infiltration and therapeutic outcomes. Front Immunol. 2020;11:629722.
doi pubmed - Han B, Gu Z, Liu Z, Ling H. Clinical characteristics and survival outcomes of infiltrating lobular carcinoma: a retrospective study of 365 cases in China. Cancer Manag Res. 2022;14:647-658.
doi pubmed - Oesterreich S, Nasrazadani A, Zou J, Carleton N, Onger T, Wright MD, Li Y, et al. Clinicopathological features and outcomes comparing patients with invasive ductal and lobular breast cancer. J Natl Cancer Inst. 2022;114(11):1511-1522.
doi pubmed - Loibl S, Volz C, Mau C, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, et al. Response and prognosis after neoadjuvant chemotherapy in 1,051 patients with infiltrating lobular breast carcinoma. Breast Cancer Res Treat. 2014;144(1):153-162.
doi pubmed - Riba LA, Russell T, Alapati A, Davis RB, James TA. Characterizing response to neoadjuvant chemotherapy in invasive lobular breast carcinoma. J Surg Res. 2019;233:436-443.
doi pubmed - Cristofanilli M, Gonzalez-Angulo A, Sneige N, Kau SW, Broglio K, Theriault RL, Valero V, et al. Invasive lobular carcinoma classic type: response to primary chemotherapy and survival outcomes. J Clin Oncol. 2005;23(1):41-48.
doi pubmed - Delpech Y, Coutant C, Hsu L, Barranger E, Iwamoto T, Barcenas CH, Hortobagyi GN, et al. Clinical benefit from neoadjuvant chemotherapy in oestrogen receptor-positive invasive ductal and lobular carcinomas. Br J Cancer. 2013;108(2):285-291.
doi pubmed - Desmedt C, Salgado R, Fornili M, Pruneri G, Van den Eynden G, Zoppoli G, Rothe F, et al. Immune infiltration in invasive lobular breast cancer. J Natl Cancer Inst. 2018;110(7):768-776.
doi pubmed - He Q, Xue S, Wa Q, He M, Feng S, Chen Z, Chen W, et al. Mining immune-related genes with prognostic value in the tumor microenvironment of breast invasive ductal carcinoma. Medicine (Baltimore). 2021;100(17):e25715.
doi pubmed - Wu R, Oshi M, Asaoka M, Yan L, Benesch MGK, Khoury T, Nagahashi M, et al. Intratumoral Tumor Infiltrating Lymphocytes (TILs) are associated with cell proliferation and better survival but not always with chemotherapy response in breast cancer. Ann Surg. 2023;278(4):587-597.
doi pubmed - McCart Reed AE, Lal S, Kutasovic JR, Wockner L, Robertson A, de Luca XM, Kalita-de Croft P, et al. LobSig is a multigene predictor of outcome in invasive lobular carcinoma. NPJ Breast Cancer. 2019;5:18.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.