| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Review
Volume 16, Number 1, February 2025, pages 1-15
MicroRNA-155 as Biomarker and Its Diagnostic Value in Breast Cancer: A Systematic Review
Bann Siang Yeoa, Wen Xuan Leea, Rozi Mahmudb, Geok Chin Tanc, Mohamed Ibrahim Abdul Wahidd, Yoke Kqueen Cheaha, e, f
aDepartment of Biomedical Science, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Selangor, Malaysia
bDepartment of Radiology, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Selangor, Malaysia
cDepartment of Pathology, Faculty of Medicine, Universiti Kebangsaan Malaysia, Wilayah Persekutuan Kuala Lumpur, Malaysia
dDepartment of Oncology, Beacon Hospital Sdn. Bhd., Petaling JayaSelangor, Malaysia
eUPM-MAKNA Cancer Research Laboratory, Institute of Bioscience, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
fCorresponding Author: Yoke Kqueen Cheah, Department of Biomedical Science, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Selangor, Malaysia
Manuscript submitted August 27, 2024, accepted October 15, 2024, published online December 31, 2024
Short title: MiR-155 as Diagnostic Breast Cancer Biomarker
doi: https://doi.org/10.14740/wjon1955
| Abstract | ▴Top |
The investigation of microRNAs (miRNAs) for the purpose of identifying biomarkers and new treatments for breast cancer has been gaining traction from scientists in recent years. Of all the miRNAs, miR-155 has been reportedly involved in breast cancer development as it regulates various cellular processes such as glucose uptake, proliferation, metastasis, and migration. Various efforts have been done towards researching miR-155 as a biomarker in breast cancer; however, the results were varied. The objective of the current systematic review is to compile and summarize information regarding miR-155 as a potential diagnostic biomarker for breast cancer. All eligible studies were found from SCOPUS and PubMed databases. Out of the 376 potential eligible records, only 26 original articles were selected for further assessment according to inclusion and exclusion criteria. The expressions of miR-155 in serum, plasma, biopsy, urine, nipple aspirate fluid, serum exosomes, and peripheral blood mononuclear cells were recorded and analyzed. Besides that, the expression of miR-155 was also correlated to clinicopathological features in breast cancer patients. The area under the curve (AUC) values from receiver operating characteristic (ROC) analysis used to evaluate diagnostic sensitivity and specificity of miR-155 as a diagnostic biomarker were also recorded. The limitations such as the small sampling size, the unemployment of internal controls for quantitative real-time polymerase chain reaction (RT-qPCR), and inconsistency of sensitivity as well as specificity values of miR-155 as a biomarker have been discussed. The present study proposed that miR-155 is a good diagnostic biomarker for breast cancer; however, further clinical research is required to assess the validity of miR-155 as a potential biomarker to translate the research outcomes into clinical practice.
Keywords: MicroRNA; MiR-155; Diagnostics; Biomarker; Breast cancer; Systematic review
| Introduction | ▴Top |
Breast cancer has been an alarming issue globally as it is the leading cause of cancer-related death in women [1]. Female breast cancer is the most commonly diagnosed cancer worldwide, with 2.3 million affected cases and 685,000 mortality cases reported in 2020 [2]. Devastatingly, of those 2.3 million affected individuals, 45.4% of those breast cancer cases were diagnosed and reported in Asia [3]. According to World Health Organization (WHO), Asia was ranked the second highest for mortality-to-incidence ratio of breast cancer (0.32) which is higher than the world’s average (0.28) [4].
The advancement of technology in the healthcare has allowed us to develop various equipment such as mammography [5], ultrasound [6], and magnetic resonance imaging (MRI) [7] for diagnostic screening and disease monitoring to aid in the early detection and prognosis of breast cancer. Mammography, which has been the gold standard for breast cancer screening, has certain limitations such as false positives and false negative findings, leading to misdiagnosis [8]. Therefore, blood-based molecular biomarkers with high diagnostic performance could be alternatives to improve and complement the current imaging techniques used for diagnosing breast cancer.
MicroRNAs (miRNAs) are short, non-coding RNA transcripts derived from the metabolic processing of longer, primary RNA transcripts encoded by miR genes [9]. Deregulated miRNA expression profiles have been implicated in various human cancers such as breast [10], lung [11], colon [12], and adult T-cell leukemia [13]. Therefore, circulating miRNAs have gained widespread attention in the field of molecular and cancer biology for its proposed practicality as novel biomarkers in many human diseases due to its stable nature [14]. Of all the identified miRNAs, miR-155 has been reported to be highly correlated with breast cancer development in women. MiR-155 maps within and is processed from an exon of a non-coding RNA transcribed from the B-cell integration cluster (BIC) located on chromosome 21. MiR-155 has been reported to play essential roles in pathological and physiological processes in the human body as it regulates the expression of other oncogenes and tumor-suppressors associated with breast cancer carcinogenesis [15].
Although miR-155 has been proposed for various diagnostic and prognostic purposes in breast cancer [16], some of these studies had contradicting outcomes. For example, Huang and co-workers (2018) reported high diagnostic efficiency and reliable clinical validity for miR-155, which demonstrated its diagnostic ability as a biomarker in breast cancer [17]. On the other hand, a study by Grimaldi et al (2020) stated that miR-155 was one of the most non-specific biomarkers for diagnostic purposes due to its poor consistent expression patterns and lack of specificity, for which the role and mechanism of miR-155 needs further verification [18]. As a result, we considered that it would be useful to conduct a systematic review on the diagnostic values of miR-155 as observed in several human samples such as serum, plasma, tissue biopsy, urine, peripheral blood mononuclear cells, and serum exosomes from breast cancer patients in order to solidify the diagnostic performance of miR-155 as a prospective biomarker for breast cancer diagnosis.
| Materials and Methods | ▴Top |
Search strategy
A systematic literature search was performed by two authors (YBS and LWX) independently on SCOPUS and PubMed databases. The following search strategies were used: (miR-155 OR microRNA-155 OR miRNA-155 OR miR155) AND breast cancer AND (diagnosis OR diagnostic OR biomarker OR biopsy OR blood OR plasma OR serum).
Quality assessment
The inclusion and exclusion criteria of the articles are as follows. Inclusion criteria were: 1) miR-155 as a diagnostic tool in blood samples of breast cancer patients, breast cancer tissue or any relevant samples collected from breast cancer patients; 2) original research articles; 3) English language; and 4) sample collection was done before patients were given treatment. Exclusion criteria were: 1) conference proceedings, letters, case reports, and reviews; 2) articles in press; 3) non-English articles; 4) methodological studies; 5) studies that do not focus on breast cancer and miR-155; and 6) full text not available. The discrepancies between the authors were resolved by consensus.
A quality assessment of selected studies was conducted by utilizing the criteria set by Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) [19]. Each item on the QUADAS-2 list was checked with answers, yes, no or unclear. Then, the scores of QUADAS-2 were recorded to determine the overall quality of the selected studies. The scores of each article were reported. This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA).
Data extraction and quality assessment
After the selection procedure, a summary was prepared by an author (YBS) using Excel spreadsheet database to include all the articles which met the inclusion criteria. Further information verification was done by the second author (LWX). For each study, the following information was extracted: publication year, sample used for investigation, cohort study, miRNA investigated in the study, method used to investigate miR-155 expression, expression of miR-155 in samples, clinicopathological features, area under the curve (AUC) of receiver operating characteristic (ROC), sensitivity, specificity, and reference.
| Results | ▴Top |
Literature search results
According to the proposed search strategy, a total of 376 articles related to the diagnostic role of miR-155 in breast cancer were found from SCOPUS and PubMed databases. After removing the duplicates, 307 records remained. Of these potential eligible records, 47 records were excluded because they were either original research articles or non-English literature publications. From the remaining 260 articles, 155 of the records were found to be irrelevant after title and abstract screening, leaving 105 potential eligible records to be screened. Of these articles, 11 records were focused on the prognostic value of miR-155 that will not be discussed in this article, 25 were methodological studies, 10 involved the use of cell lines or animal models, and 34 articles were found lacking relevant information pertaining to miR-155. The consensus for full-text acceptance between two authors was 75.8% (n = 25/33). For disputed articles (n = 8), one article was included whereas the other of the articles were rejected after discussion between two authors. As a result, this systematic review was written based on 26 eligible studies which discussed the diagnostic value of miR-155 in serum, plasma or biopsy samples from breast cancer patients. Furthermore, we also included studies which investigated the expression of miR-155 in urine samples, nipple aspirate fluid, serum exosomes, and peripheral blood mononuclear cells of breast cancer patients. Figure 1 shows the results of the literature search.
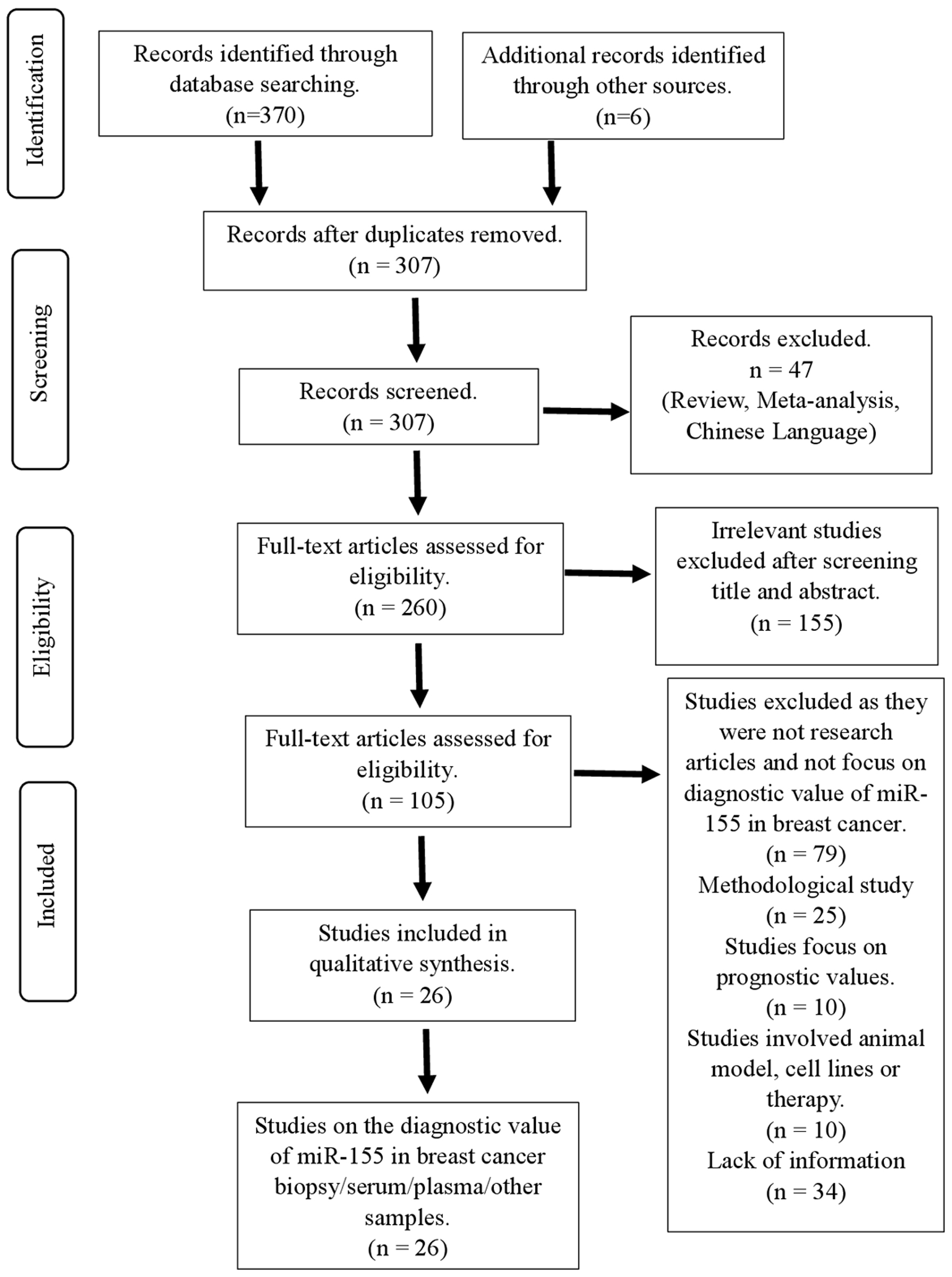 Click for large image | Figure 1. Flowchart showing strategy searches and selection processes for the systematic review. |
The findings of each study are detailed in Table 1 [17, 20-37] and Table 2 [27, 34, 38-44]. In this systematic review, we have selected 26 original articles that were published between 2014 and 2024 and had a sample size ranging from 14 to 290 samples, amounting to 2,901 samples in total. Twenty-two included studies assessed miR-155 in a shortlist of two or more deregulated miRNAs, while four articles were focused solely on miR-155 as a biomarker. All 26 studies utilized quantitative real-time polymerase chain reaction (RT-qPCR) as the method to detect miRNA expression. Pertaining to the types of samples used for miR-155 expression, 10 studies used serum samples, seven articles used plasma samples, three studies used breast cancer tissue biopsies, one study investigated serum exosomes, one study used nipple aspirate fluid, one study used peripheral blood mononuclear cells of breast cancer patients, and the remaining two studies investigated multiple biological samples such as midstream specimen of urine and serum as well as plasma and tissue biopsy, respectively. Furthermore, 17 out of 26 articles evaluated the biomarker potential of miR-155 using ROC curve analysis. The AUC values were calculated to evaluate the diagnostic accuracy of miR-155 as a diagnostic biomarker. The specificity and sensitivity values of miR-155 were also reported.
 Click to view | Table 1. Comprehensive Characteristics of Selected Studies, Association of miR-155 Expression in Serum or Plasma and Clinicopathological Features of Breast Cancer Patients |
 Click to view | Table 2. Comprehensive Characteristics of Selected Studies, Association of miR-155 Expression of Biological Samples and Clinicopathological Features in Breast Cancer Patients |
Quality assessment
The quality of the 26 studies was evaluated using QUADAS-2, and the majority of the selected articles scored more than 6 out of 9. In addition, most of the selected articles were ranked as low risk of bias with low risk in applicability. Only one study was rated 3 out of 9 due to unclear exclusion criteria of patient recruitment, low number of patient sampling as well as unclear internal reference used for RT-qPCR. Despite that, six studies (23%) were found to have high risk of biased patient selection, while unclear reference standard was found in 19% of the selected studies. As a result, significant bias was not presented. The detailed information of QUADAS-2 assessment is presented in Tables 3 and 4 [17, 20-44] as well as Figures 2 and 3.
 Click to view | Table 3. Quality Assessment for All Selected Articles Based on Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) and the Score of Each Section |
 Click to view | Table 4. Risk of Bias and Applicability of the Methodology of Each Selected Study |
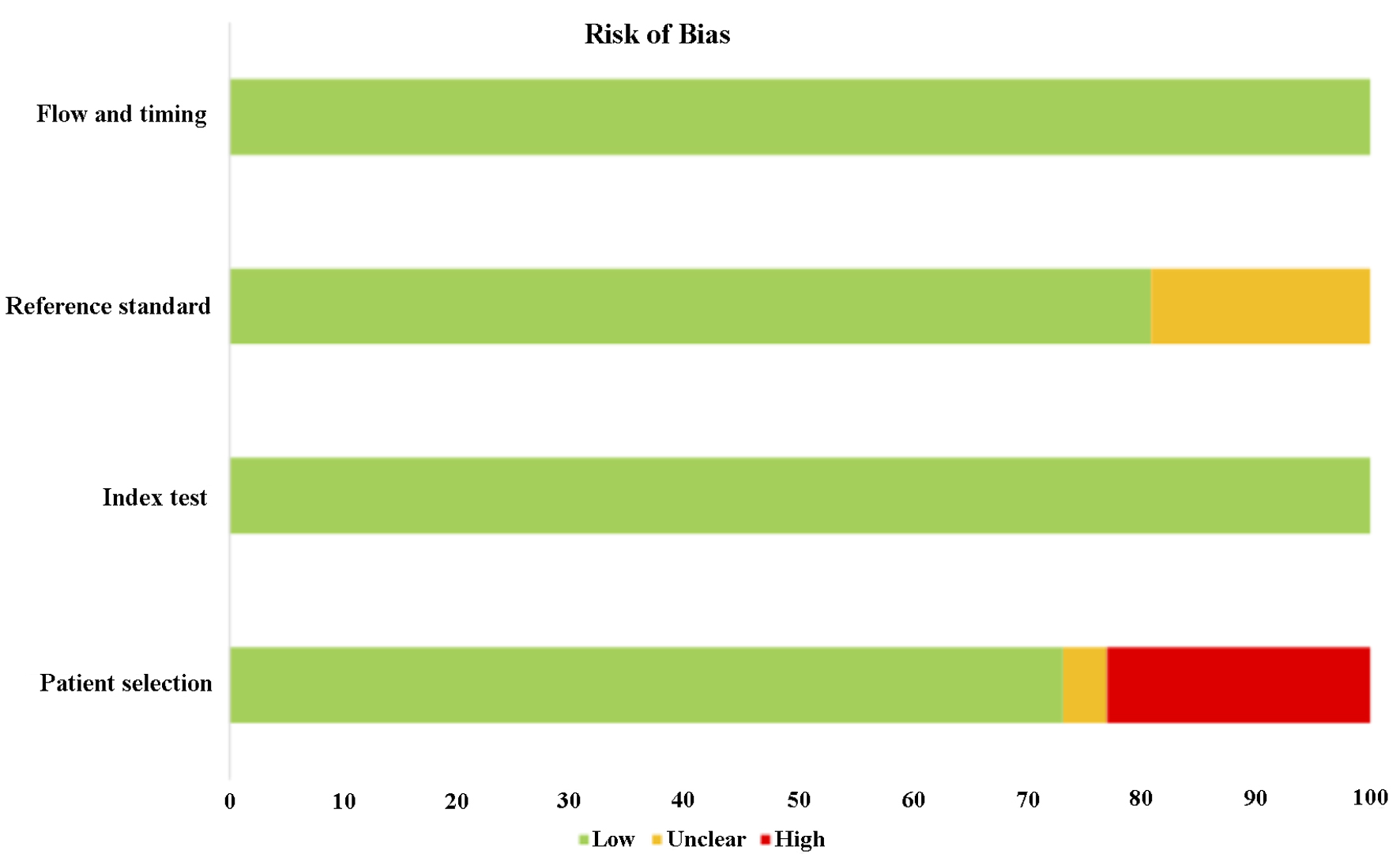 Click for large image | Figure 2. The risk of bias of the methodology of the selected studies. All the studies were ranked low risk of bias in flow and timing and index test. However, 23% of studies were found to have high risk of bias in patient’s selection, while 19% of the studies were ranked unclear in reference standard. |
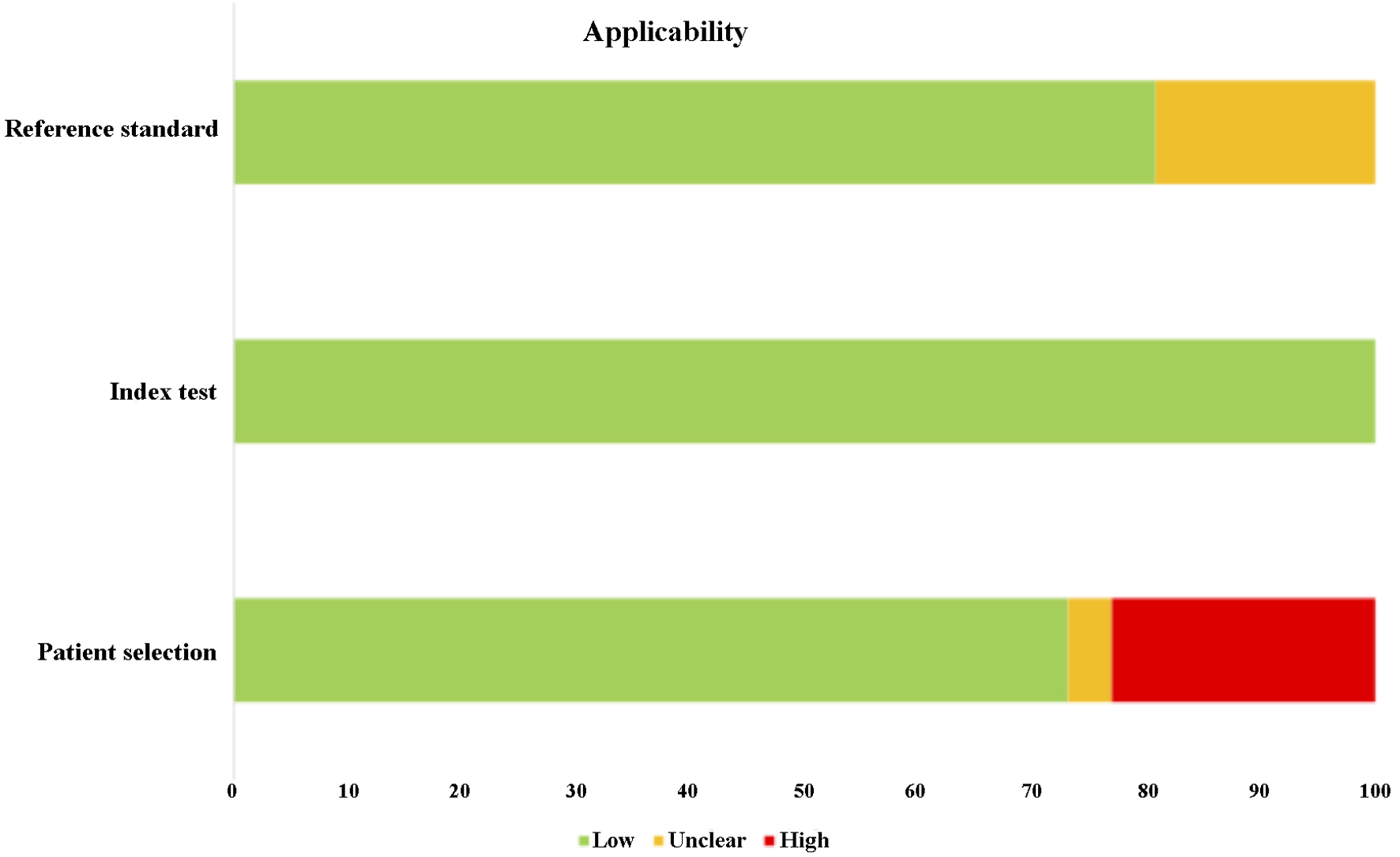 Click for large image | Figure 3. The applicability of the methodology of the selected studies. All the studies were ranked low risk of error in index test. However, 23% of studies were found to have high risk of errors in patient’s selection, while 19% of the studies were ranked unclear in reference standard. |
Diagnostic value of miR-155
All articles focused on miR-155 as a diagnostic biomarker for both invasive and non-invasive breast cancers were investigated. Among the total 26 selected articles, 14 research articles correlated miR-155 to the clinicopathological changes (tumor size, stage of cancer, grade of cancer, involvement of lymph node, metastasis, and molecular type of tumor) of breast cancer.
Expression of miR-155 in serum/plasma
Of all the articles, 19 of them involved the investigation of miR-155 expression in breast cancer patients’ serum or plasma [17, 20-37], 15 articles have demonstrated upregulation of miR-155 in breast cancer patients’ serum or plasma [17, 20-33], one study showed no significant difference in the expression of miR-155 in serum of healthy individuals and breast cancer patients [34], and three remaining articles showed downregulation of miR-155 expression [35-37].
Among the selected articles, three studies investigated miR-155 expression in invasive breast cancer patients’ serum or plasma. Huang et al (2018) and Anwar et al (2020) demonstrated significant upregulation of miR-155 in serum and plasma respectively from invasive breast cancer patients compared to healthy controls [17, 20]. On the other hand, Jurkovicova et al (2017) showed that miR-155 was downregulated by 0.71-fold in plasma extracted from invasive breast cancer in respect to healthy controls [35].
Three studies examined the expression of miR-155 in early breast cancer patients. Itani et al (2021), Canatan et al (2021), and Mohamed et al (2022) found that miR-155 was significantly upregulated in the samples from early breast cancer patients as compared to healthy subjects [21-23]. Besides that, two articles focused on miR-155 expression in primary breast cancer patients have shown conflicting results. In a study by Swellam et al (2019), miR-155 was significantly expressed in serum samples of primary breast cancer patients compared to those that are benign [24]. However, Erbes et al (2015) disclosed that there was no significant difference in miR-155 expression in the serum between primary breast cancer and healthy individuals [34].
In addition, a few authors have put their attention into investigating miR-155 expression in the serum or plasma of specific types of breast cancers. A study by Aksan et al (2020) examined miRNA expression in patients with idiopathic granulomatous mastitis (IGM), revealing that miR-155 expression in IGM patients was downregulated by 0.73-fold compared to healthy controls [36], whereas Shaheen et al (2019) mentioned that miR-155 expression was upregulated in triple negative breast cancer (TNBC) plasma samples by 26.21-fold, compared to non-TNBC plasma samples (9.57-fold) [25]. Similar results were reported by Kumar et al (2023) who discovered the elevated miR-155 expression with a fold change of 7.26 in TNBC patients’ serum sample compared to serum samples from healthy individuals [26].
Expression of miR-155 in biopsy, urine, serum exosomes, nipple aspirate fluid, and peripheral blood mononuclear cells
A few studies have also investigated the miR-155 expression in other biological samples such as breast cancer tissue biopsy, urine, serum exosomes, nipple aspirate fluid, and peripheral blood mononuclear cells. Soleimanpour et al (2019) and Bitaraf et al (2020) mentioned that miR-155 was significantly upregulated in invasive breast cancer tumor tissues [27, 38]. Similar results were also reported by Nassar et al (2014) and Kolesnikov et al (2016), who observed upregulation of miR-155 in tissues of breast cancer patients [39, 40]. Erbes et al (2015) found that urine samples can also be utilized for breast cancer diagnosis and observed that miR-155 was overexpressed in the urine of breast cancer patients [34]. Asgeri et al (2022) have proved that serum exosomes could be utilized as testing samples for breast cancer diagnosis as expression of miR-155 in serum exosomes from breast cancer patients was higher compared to controls [41]. Moreover, recent studies by Raeisi et al (2020) and Iranparast et al (2022) reported miR-155 upregulation in peripheral blood mononuclear cells of newly diagnosed breast cancer patients [42, 43]. However, unlike other studies, Qin et al (2017) mentioned that there were no significant differences in miR-155 expression detected in nipple aspirate fluid between breast cancer patients and healthy controls [44].
MiR-155 as a potential diagnostic marker for breast cancer
Of the 26 selected articles, 17 research articles evaluated the diagnostic value of miR-155 using AUC values calculated from ROC. AUC values closer to 1 reflect a more substantial difference between breast cancer samples and normal samples. Ranges of AUC values between 0.5 and 0.7 will be considered as low diagnostic accuracy, whereas moderate diagnostic accuracy falls within the 0.7 to 0.9 range; AUC values higher than 0.9 will indicate high diagnostic accuracy [28]. In addition, the sensitivity and specificity values of miR-155 were reported in only 13 selected articles.
Overall, the AUC values of the ROC curve for miR-155 expressed in serum or plasma samples from the selected articles ranged from 0.692 to 0.995. Majority of the selected studies had AUC values that reflect the moderate diagnostic accuracy category. For example, Guo et al (2016) and Mojahed et al (2020) who assessed the expression of miR-155 solely in serum reported AUC values of 0.879 and 0.890, respectively, indicating moderate diagnostic accuracy [28, 29]. High AUC values, 0.944 and 0.995, were reported by Mohamed et al (2022) and Swellam et al (2019), respectively, where both authors investigated more than two miRNAs [23, 30]. On the other hand, the AUC value reported by Zhang et al (2016), 0.692, was lower than 0.7, reflecting low diagnostic accuracy [31].
Sensitivity and specificity values for detecting miR-155 expression in serum and plasma samples had sensitivity values ranging from 66% to 100% and specificity values ranging from 51.02% to 97.10%. Studies by Shaheen et al (2019) and Han et al (2016) both reported 100.00% sensitivity in detecting miR-155 of serum samples, despite the specificity being only 73.53% and 51.02%, respectively [25, 32]. On the other hand, 91% and 88.89% specificity in identifying miR-155 were reported by Anwar et al (2020) and Mojahed et al (2020), despite the sensitivity of using miR-155 as biomarker being reported at 70% and 77.78%, respectively [20, 29]. A study by Swellam et al (2019) reported high sensitivity (95.00%) and high specificity (97.10%) in detecting miR-155 in serum [24]. In addition, low sensitivity (66%) and low specificity values (68.9%) were also reported by Zhang et al (2016) [31].
Besides that, ROC analysis was conducted in studies which employed other biological samples for the investigation of miR-155 as a biomarker for breast cancer. High diagnostic accuracy with AUC values of 0.83 and 0.941 was reported by Soleimanpour et al (2018) and Raeisi et al (2020), respectively from breast cancer tissue biopsy samples [27, 42]. In addition, Raeisi et al (2020) also reported high sensitivity (84.4%) and specificity (91.1%) of using miR-155 as a biomarker in breast cancer tissue biopsy [42]. Moderate diagnostic accuracy, with an AUC value of 0.87, was reported by Kolesnikov et al (2016), who investigated the expression of miR-155 in tissue biopsy from TNBC patients [40].
Correlation of miR-155 expression and clinicopathological features in breast cancer patients
Out of the 26 selected articles, 15 of them associated miR-155 expression in serum, plasma or tissue biopsy samples with the clinicopathological features in breast cancer patients. Only Iranparast et al (2022) investigated the association of breast cancer clinicopathological features and miR-155 expression in peripheral blood mononuclear cells [43]. Han et al (2016), Bitaraf et al (2019), and Nassar et al (2014) reported the correlation of miR-155 expression with clinicopathological changes in breast cancer from tissue biopsies, while the remaining articles reported these correlations using serum or plasma samples [32, 38, 39]. In general, different findings have been reported.
Among the chosen articles, five authors investigated the correlation between breast cancer tumor sizes and the changes of miR-155 levels. Anwar et al (2020), Mohamed et al (2022), and Mojahed et al (2020) stated that miR-155 expression was high when the tumor size was larger than 5 cm [20, 23, 29]. On the other hand, Shaheen et al (2019) revealed that the expression of miR-155 decreased when the breast cancer tumor size increased [25]. Besides that, the study done by Anwar et al (2020) reported that miR-155 expression was high in grade III breast cancer which aligned with the findings of Mojahed et al (2020), Swellam et al (2019), and Hagrass et al (2014) [20, 29, 30, 33]. Contrastingly, Shaheen et al (2019) observed decreased miR-155 expression in stage III and IV of breast cancer [25].
In addition, the association between miR-155 expression and patients’ age, lymph node involvement, and metastasis status was investigated. Anwar et al (2020) observed that breast cancer patients who are 40 years and above had higher levels of miR-155 in their plasma [20]. Similar findings were reported by Nassar et al (2014), who studied the miR-155 expression in breast cancer tumors [39]. In contrast, Soleimanpour et al (2018) reported no correlation between breast cancer patients’ age and miR-155 expression in plasma [27]. In the case of lymph node involvement, Shaheen et al (2019) discovered that miR-155 levels were decreased in advanced stages of breast cancer, such as N2 and N3 stages [25]. Furthermore, two studies have reported significant increases in miR-155 expression in metastatic breast cancer and breast cancer patients with lymph node metastasis [33, 37]. However, Shaheen et al (2019) reported low levels of miR-155 in case of metastasis in advanced stages of TNBC [25].
In three out of 15 articles that investigated the correlation of miR-155 expression and molecular classification of breast cancer, Jurkovicova et al (2017) reported that increased miR-155 expression was associated with estrogen receptor (ER)-positive and progesterone receptor (PR)-positive as well as high level of Ki-67 breast cancer [35]. In contrast, Mohamed et al (2022) and Nassar et al (2014) demonstrated high levels of miR-155 in PR-negative breast cancer and human epidermal growth factor receptor 2 (HER2)-positive breast cancer [23, 39].
| Discussion | ▴Top |
In the pursuit of diagnostic biomarkers for various human diseases, researchers have proposed miRNAs as an option due to their involvement in oncological development and metastasis in breast cancer. MiRNAs play pivotal roles in the post-transcriptional regulation of gene expression in different tissues and stages of cellular development via highly specific interactions and complex regulatory networks [45]. Many studies have shown significant differential miRNA expression profiles in breast cancer tissues compared to those of non-cancerous tissue [17, 24, 33]. In addition, numerous studies have been conducted to assess the potential use of circulating miRNAs found in human serum and plasma as diagnostic biomarkers for breast cancer [22, 26, 29]. Of all the miRNAs investigated, the expression of miR-155 has been widely reported in breast cancer research. To date, many researchers have demonstrated an overexpression of miR-155 in the serum and plasma of breast cancer patients. MiR-155 was shown to be involved in breast cancer proliferation and metastasis, breast cancer cell migration, as well as the regulation of metabolism, especially that of glucose uptake and glycolysis [46-48]. The overexpression of miR-155 in breast cancer tissues as well as in liquid biopsy samples, such as serum or plasma, has solidified the potential of miR-155 as a breast cancer biomarker. These studies highlight the importance of such data to stimulate the development of breast cancer diagnostic kits and targeted anti-tumor therapies that are currently not available in clinical settings and the commercial market. Hence, in this systematic review, we have analyzed the diagnostic value of miR-155 in breast cancer in order to provide useful insights by consolidating data from various studies involving the expression as well as the diagnostic value of miR-155 in various human samples such as serum, plasma, breast cancer tissue biopsies, urine, peripheral blood mononuclear cells, and serum exosomes.
We have selected 26 original articles that demonstrated the potential diagnostic values of miR-155 in breast cancer from 376 identifiable records. The outcomes of all the studies were different, with varying sensitivity and specificity values of miR-155, even though miR-155 was found to be overexpressed in majority of the studies. Generally, all authors discussed the correlation of miR-155 expression to the likelihood of breast cancer development as well as clinicopathological parameters (tumor size, stage of cancer, grade of cancer, involvement of lymph node, metastasis, and molecular subtype of the tumor) of breast cancer. One of the major challenges in breast cancer diagnosis, prognosis, and treatment is the inherent heterogeneity of breast cancer tumors [49]. Differential expression of miR-155 in various molecular subtypes of breast cancer would allow refinement of breast cancer classification for the purpose of improved diagnosis and guide cancer treatment decisions [50]. As seen from studies that correlated molecular subtypes with miR-155, upregulated miR-155 expression is reported to be associated with ER-, PR- and HER-2 positive breast cancer [23, 24]. However, conflicting results were observed by Mohamed et al (2022) where in PR-negative breast cancer, high miR-155 expression was instead reported, highlighting the need for further research regarding this correlation [23].
The majority of the findings presented the upregulation of miR-155 in the serum, plasma as well as tissue biopsy samples collected from breast cancer patients. Similar results were also discovered in urine [34], serum exosomes [41], and peripheral blood mononuclear cells [42, 43]. In contrast, three studies have shown the downregulation of miR-155 levels in plasma or serum samples from breast cancer patients [35-37], while no significant findings were reported by Erbes et al (2015) when serum samples from breast cancer patients were tested [34]. Contradictory results were observed due to the varying demographic factors among all the selected studies as well as the investigated breast cancer subtypes. In addition, we discovered that in some of the selected studies, the number of subjects involved in the research was insufficient to represent the population size of the geographical location where the study was conducted. For instance, Soleimanpour et al (2018) recruited 30 breast cancer patients while Mojahed et al (2020) had 36 breast cancer patients in their studies [27, 29]. Besides that, the number of healthy controls recruited in a few other studies was less than the number of breast cancer patients involved. The number of breast cancer patients involved in Jurkovicova et al (2017) study was at least four times (128 patients) more than the number of healthy individuals recruited (28 controls) [35]. Furthermore, although breast cancer tissue biopsies were sampled in most of the chosen studies, the tissue samples were mainly used for the purpose of clinicopathological evaluation. Only four out of the 26 selected articles presented the expression of miR-155 in breast cancer biopsy [27, 38-40]. Despite this, we still propose miR-155 to be utilized as a breast cancer diagnostic biomarker due to the high expression of miR-155 as it influences breast cancer progression for instances cell proliferation and survival, cell migration and invasion, cell apoptosis as well as drug resistance [27]. We suggest that the future study should consider larger group of research subjects which represents the population where the study is conducted.
A few studies have put their attention into investigating other human bodily fluids such as urine, peripheral blood mononuclear cells, nipple aspirate fluid, and serum exosomes. Results have shown that miR-155 could be a potential candidate as breast cancer biomarker in the aforementioned samples; however, further investigation should be conducted to study the diagnostic performance and reliability of using these samples for diagnosing breast cancer.
ROC analysis was used to evaluate the diagnostic value of miR-155. Even though all the selected articles conducted similar experiments in reporting miR-155 expression in various human samples, only 17 out of 26 articles presented the AUC values, which are sensitivity and specificity values used to evaluate miR-155 as a diagnostic biomarker. However, the results of these studies are varied. There were two articles that reported the AUC values but not the sensitivity and specificity values of miR-155 [21, 40]. Among the articles which reported AUC values, the majority of them had AUC values that scored above 0.83, while the lowest recorded AUC value was 0.692. In addition, inconsistent results of sensitivity and specificity values for miR-155 were observed among the selected studies. High sensitivity of miR-155 was found in studies by Shaheen et al (2019) (100%), Swellam et al (2019) (95%), and Han et al (2016) (100%) [25, 30, 32], while the sensitivity of miR-155 reported by Zhang et al (2016) was only 66% [31]. Similarly, the specificity of miR-155 as a biomarker varied where low specificity, 51.02%, was reported by Han et al (2016) [32], whereas 91% and 90% were reported by Anwar et al (2022) and Mohamed et al (2022), respectively [20, 23]. The inconsistency of the results could be due to the different ranges in sampling sizes, as some of the sampling sizes were not the best representation of the population within the geographical location. Furthermore, the employment of endogenous controls such as RNU6-2 [36], miR-181 [22], and miR-16 [25, 34] in normalizing the expression level of miR-155 could be helpful in standardizing the outcomes of sensitivity and specificity values of miR-155 during the ROC analysis. Nevertheless, the details of internal controls were not clearly mentioned in some of the selected articles, hence the reference standard was unclear. Instead of solely analyzing the diagnostic value of miR-155, there were studies that presented the ROC analysis as a combination panel of miRNAs that includes miR-155 in a shortlist of a few other miRNAs. For instance, an AUC value of 1.0 was reported by Mohamed et al (2022) from the miRNA combination panel of miR-155, miR-10b, miR-34a, and miR-373 [23]. As a result, the signature expression of a series of miRNAs could be an alternative or even a better indication in justifying the breast cancer diagnosis based on the AUC values stated in the selected articles.
| Conclusion and Recommendations | ▴Top |
Conclusively, the results from our systematic review show that miR-155 has the potential to be a good diagnostic biomarker for breast cancer. The expressions of miR-155 in breast cancer were reported to be upregulated in samples from breast cancer patients from most studies despite them being conducted at different geographic locations with different patient demography. The results from the present study could be conducive for future scientific efforts needed to instigate the development of targeted anti-tumor therapies and breast cancer diagnostic kits using blood-based biomarkers to complement the current breast cancer screening options and treatment regimens. In spite of everything, further research still needs to be conducted on a larger group of subjects in multi-institutional studies to elucidate the role and mechanism of miR-155 as a reliable biomarker for breast cancer in order to translate subsequent research outcomes into clinical practice. Given the progress seen from miRNA-based diagnostic kits in other human cancers such as lung [51], pancreatic [52], thyroid [53], and gastric cancer [54], it would not be far-fetched to hope for the same outcomes for breast cancer. However, inter- and intra-tumor heterogeneity of breast cancer tumors still presents significant challenges in breast cancer classification and treatment [49]. Refinement in breast cancer stratification using miRNA-based diagnostic kits could complement current diagnosis techniques and encourage advancements in personalized precision treatment of breast cancer, with the aim of reducing the clinical burden of breast cancer patients.
Acknowledgments
We would like to express our gratitude to the support of funding, Technology Development Fund 1, TeD(1)/2023/5450832 (TDF06221585), from Ministry of Science, Technology and Innovation (MOSTI) of Malaysia.
Financial Disclosure
This work was funded by the Ministry of Science, Technology and Innovation (MOSTI) of Malaysia under the Development Fund 1 (TeD1, TDF06221585).
Conflict of Interest
The authors declare that there is no competing financial interests or personal relationships that can appear to influence the work presented in this article.
Author Contributions
BSY: conceived study idea, data search, statistical analysis, designed tables and figures, and drafted the manuscript. WXL: data search, drafted the manuscript. RM: review and revision of the manuscript draft. GCT: review and revision of the manuscript draft. MIAW: review and revision of the manuscript draft. YKC: review and revision of the manuscript draft.
Data Availability
The authors declare that data supporting the findings of this study are available within the article
| References | ▴Top |
- Sokolova A, Johnstone KJ, McCart Reed AE, Simpson PT, Lakhani SR. Hereditary breast cancer: syndromes, tumour pathology and molecular testing. Histopathology. 2023;82(1):70-82.
doi pubmed - Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, Vignat J, et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast. 2022;66:15-23.
doi pubmed - Lim YX, Lim ZL, Ho PJ, Li J. Breast cancer in Asia: incidence, mortality, early detection, mammography programs, and risk-based screening initiatives. Cancers (Basel). 2022;14(17):4218.
doi pubmed - World Health Organisation (WHO). Estimated age-standardized incidence rates (world) in 2020, Breast, Female, All Ages, Asia. 2020.
- Bernardi D, Macaskill P, Pellegrini M, Valentini M, Fanto C, Ostillio L, Tuttobene P, et al. Breast cancer screening with tomosynthesis (3D mammography) with acquired or synthetic 2D mammography compared with 2D mammography alone (STORM-2): a population-based prospective study. Lancet Oncol. 2016;17(8):1105-1113.
doi pubmed - Geisel J, Raghu M, Hooley R. The role of ultrasound in breast cancer screening: the case for and against ultrasound. Semin Ultrasound CT MR. 2018;39(1):25-34.
doi pubmed - Morrow M, Waters J, Morris E. MRI for breast cancer screening, diagnosis, and treatment. Lancet. 2011;378(9805):1804-1811.
doi pubmed - Winters S, Martin C, Murphy D, Shokar NK. Breast cancer epidemiology, prevention, and screening. Prog Mol Biol Transl Sci. 2017;151:1-32.
doi pubmed - Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004.
doi pubmed - Medimegh I, Omrane I, Privat M, Uhrhummer N, Ayari H, Belaiba F, Benayed F, et al. MicroRNAs expression in triple negative vs non triple negative breast cancer in Tunisia: interaction with clinical outcome. PLoS One. 2014;9(11):e111877.
doi pubmed - Souza CP, Cinegaglia NC, Felix TF, Evangelista AF, Oliveira RA, Hasimoto EN, Cataneo DC, et al. Deregulated microRNAs Are Associated with Patient Survival and Predicted to Target Genes That Modulate Lung Cancer Signaling Pathways. Cancers (Basel). 2020;12(9):2711.
doi pubmed - Ahadi A. The significance of microRNA deregulation in colorectal cancer development and the clinical uses as a diagnostic and prognostic biomarker and therapeutic agent. Noncoding RNA Res. 2020;5(3):125-134.
doi pubmed - Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, et al. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell. 2012;21(1):121-135.
doi pubmed - Sandau US, Wiedrick JT, McFarland TJ, Galasko DR, Fanning Z, Quinn JF, Saugstad JA. Analysis of the longitudinal stability of human plasma miRNAs and implications for disease biomarkers. Sci Rep. 2024;14(1):2148.
doi pubmed - Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792(6):497-505.
doi pubmed - Liu K, Zhao K, Wang L, Sun E. Prognostic value of microRNA-155 in human carcinomas: an updated meta-analysis. Clin Chim Acta. 2018;479:171-180.
doi pubmed - Huang SK, Luo Q, Peng H, Li J, Zhao M, Wang J, Gu YY, et al. A panel of serum noncoding RNAs for the diagnosis and monitoring of response to therapy in patients with breast cancer. Med Sci Monit. 2018;24:2476-2488.
doi pubmed - Grimaldi AM, Nuzzo S, Condorelli G, Salvatore M, Incoronato M. Prognostic and clinicopathological significance of MiR-155 in breast cancer: a systematic review. Int J Mol Sci. 2020;21(16):5834.
doi pubmed - Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529-536.
doi pubmed - Anwar SL, Tanjung DS, Fitria MS, Kartika AI, Sari DNI, Rakhmina D, Wardana T, et al. Dynamic changes of circulating Mir-155 expression and the potential application as a non-invasive biomarker in breast cancer. Asian Pac J Cancer Prev. 2020;21(2):491-497.
doi pubmed - Itani MM, Nassar FJ, Tfayli AH, Talhouk RS, Chamandi GK, Itani ARS, Makoukji J, et al. A signature of four circulating microRNAs as potential biomarkers for diagnosing early-stage breast cancer. Int J Mol Sci. 2021;22(11):6121.
doi pubmed - Canatan D, Sonmez Y, Yilmaz O, Cim A, Coskun HS, Sezgin Goksu S, Ucar S, et al. MicroRNAs as biomarkers for breast cancer. Acta Biomed. 2021;92(2):e2021028.
doi pubmed - Mohamed AA, Allam AE, Aref AM, Mahmoud MO, Eldesoky NA, Fawazy N, Sakr Y, et al. Evaluation of expressed microRNAs as prospective biomarkers for detection of breast cancer. Diagnostics (Basel). 2022;12(4):789.
doi pubmed - Swellam M, Zahran RFK, Abo El-Sadat Taha H, El-Khazragy N, Abdel-Malak C. Role of some circulating MiRNAs on breast cancer diagnosis. Arch Physiol Biochem. 2019;125(5):456-464.
doi pubmed - Shaheen J, Shahid S, Shahzadi S, Ahktar MW, Sadaf S. Identification of circulating miRNAs as non-invasive biomarkers of triple negative breast cancer in the population of Pakistan. Pakistan J. Zool. 2019;51:1113-1121.
- Kumar V, Gautam M, Chaudhary A, Chaurasia B. Impact of three miRNA signature as potential diagnostic marker for triple negative breast cancer patients. Sci Rep. 2023;13(1):21643.
doi pubmed - Soleimanpour E, Babaei E, Hosseinpour-Feizi MA, Montazeri V. Circulating miR-21 and miR-155 as potential noninvasive biomarkers in Iranian Azeri patients with breast carcinoma. J Cancer Res Ther. 2019;15(5):1092-1097.
doi pubmed - Guo J, Jiang W, Xu X, Zheng X. Serum microRNA-155 in early diagnosis and prognosis of breast cancer. Int J Clin Exp Med. 2016;9(6):10289-10296.
- Hosseini Mojahed F, Aalami AH, Pouresmaeil V, Amirabadi A, Qasemi Rad M, Sahebkar A. Clinical evaluation of the diagnostic role of microRNA-155 in breast cancer. Int J Genomics. 2020;2020:9514831.
doi pubmed - Swellam M, Ramadan A, El-Hussieny EA, Bakr NM, Hassan NM, Sobeih ME, EzzElArab LR. Clinical significance of blood-based miRNAs as diagnostic and prognostic nucleic acid markers in breast cancer: Comparative to conventional tumor markers. J Cell Biochem. 2019;120(8):12321-12330.
doi pubmed - Zhang J, Jiang C, Shi X, Yu H, Lin H, Peng Y. Diagnostic value of circulating miR-155, miR-21, and miR-10b as promising biomarkers in human breast cancer. Int J Exp Pathol. 2016;9(10):10258-10265.
- Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92(2):55-66.
doi pubmed - Hagrass HA, Sharaf S, Pasha HF, Tantawy EA, Mohamed RH, Kassem R. Circulating microRNAs - a new horizon in molecular diagnosis of breast cancer. Genes Cancer. 2015;6(5-6):281-287.
doi pubmed - Erbes T, Hirschfeld M, Rucker G, Jaeger M, Boas J, Iborra S, Mayer S, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;15:193.
doi pubmed - Jurkovicova D, Smolkova B, Magyerkova M, Sestakova Z, Kajabova VH, Kulcsar L, Zmetakova I, et al. Down-regulation of traditional oncomiRs in plasma of breast cancer patients. Oncotarget. 2017;8(44):77369-77384.
doi pubmed - Aksan H, Kundaktepe BP, Sayili U, Velidedeoglu M, Simsek G, Koksal S, Gelisgen R, et al. Circulating miR-155, let-7c, miR-21, and PTEN levels in differential diagnosis and prognosis of idiopathic granulomatous mastitis and breast cancer. Biofactors. 2020;46(6):955-962.
doi pubmed - Mohamed MA, Abdallah ZF, Nassar HR, Hilal A, ElDesouki H, Said M, Elsalam IA. Deregulated expression of candidate MicroRNAs and BRCA mutations frequency in breast cancer patients. Egypt J Chem. 2022;65(6):23-35.
- Bitaraf A, Babashah S, Garshasbi M. Aberrant expression of a five-microRNA signature in breast carcinoma as a promising biomarker for diagnosis. J Clin Lab Anal. 2020;34(2):e23063.
doi pubmed - Nassar FJ, El Sabban M, Zgheib NK, Tfayli A, Boulos F, Jabbour M, El Saghir NS, et al. miRNA as potential biomarkers of breast cancer in the Lebanese population and in young women: a pilot study. PLoS One. 2014;9(9):e107566.
doi pubmed - Kolesnikov NN, Veryaskina YA, Titov SE, Rodionov VV, Gening TP, Abakumova TV, Kometova VV, et al. Expression of microRNAs in molecular genetic breast cancer subtypes. Cancer Treat Res Commun. 2019;20:100026.
doi pubmed - Asgari R, Rezaie J. Differential expression of serum exosomal miRNAs in breast cancer patients and healthy controls. Adv Pharm Bull. 2022;12(4):858-862.
doi pubmed - Raeisi F, Mahmoudi E, Dehghani-Samani M, Hosseini SSE, Ghahfarrokhi AM, Arshi A, Forghanparast K, et al. Differential expression profile of miR-27b, miR-29a, and miR-155 in chronic lymphocytic leukemia and breast cancer patients. Mol Ther Oncolytics. 2020;16:230-237.
doi pubmed - Iranparast S, Tahmasebi-Birgani M, Motamedfar A, Amari A, Ghafourian M. Altered expression levels of microRNA-155 and SOCS-1 in peripheral blood mononuclear cells of newly diagnosed breast cancer patients. Iran J Allergy Asthma Immunol. 2022;21(1):12-19.
doi pubmed - Qin WY, Zhang K, Sauter ER. Exosomal miRNAs in nipple aspirate fluid and breast cancer. Transl Cancer Res. 2017;6(8):S1304-S1310.
- Loh HY, Norman BP, Lai KS, Rahman N, Alitheen NBM, Osman MA. The regulatory role of microRNAs in breast cancer. Int J Mol Sci. 2019;20(19):4940.
doi pubmed - Liu JH, Yang Y, Song Q, Li JB. MicroRNA-155regulates the proliferation and metastasis of human breast cancers by targeting MAPK7. J BUON. 2019;24(3):1075-1080.
pubmed - Zhang W, Chen CJ, Guo GL. MiR-155 promotes the proliferation and migration of breast cancer cells via targeting SOCS1 and MMP16. Eur Rev Med Pharmacol Sci. 2018;22(21):7323-7332.
doi pubmed - Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim J, Yoo HJ, et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. 2018;37(22):2982-2991.
doi pubmed - Guo L, Kong D, Liu J, Zhan L, Luo L, Zheng W, Zheng Q, et al. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp Hematol Oncol. 2023;12(1):3.
doi pubmed - Subramanian K, Sinha R. Functions of differentially regulated miRNAs in breast cancer progression: potential markers for early detection and candidates for therapy. Biomedicines. 2024;12(3):691.
doi pubmed - Rajakumar T, Horos R, Kittner P, Kahraman M, Sikosek T, Hinkfoth F, Tikk K, et al. Brief report: a blood-based microRNA complementary diagnostic predicts immunotherapy efficacy in Advanced-Sta ge NSCLC with high programmed death-ligand 1 express ion. JTO Clin Res Rep. 2022;3(8):100369.
doi pubmed - Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114-127.
pubmed - Finkelstein SD, Sistrunk JW, Malchoff C, Thompson DV, Kumar G, Timmaraju VA, Repko B, et al. A retrospective evaluation of the diagnostic performance of an interdependent pairwise MicroRNA expression analysis with a mutation panel in indeterminate thyroid nodules. Thyroid. 2022;32(11):1362-1371.
doi pubmed - So JBY, Kapoor R, Zhu F, Koh C, Zhou L, Zou R, Tang YC, et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut. 2021;70(5):829-837.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.