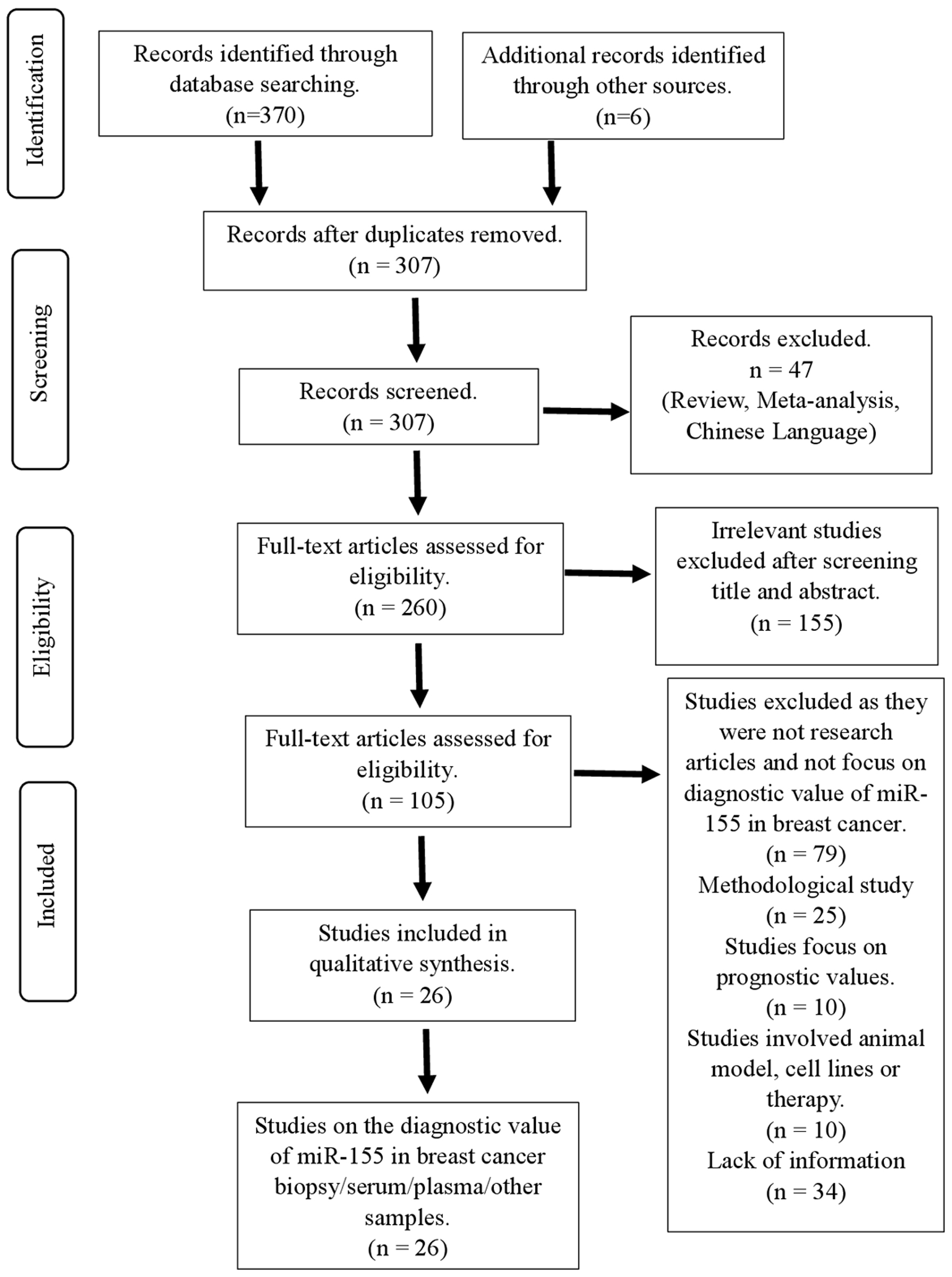
Figure 1. Flowchart showing strategy searches and selection processes for the systematic review.
| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Review
Volume 16, Number 1, February 2025, pages 1-15
MicroRNA-155 as Biomarker and Its Diagnostic Value in Breast Cancer: A Systematic Review
Figures

Tables
| No. | Year | Sample used | Cohort study | MiRNA | Method | miR-155 expression | Clinicopathological changes | Area under curve of ROC | Sensitivity, % | Specificity, % | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BC: breast cancer; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2; N/A: not applicable; PR: progesterone receptor; ROC: receiver operating characteristic; RT-qPCR: qualitative real-time polymerase chain reaction. | |||||||||||
| 1 | 2015 | Serum | 120 breast cancer patients; 50 healthy controls | miR-10b, miR-34a, miR-155, miR-195, and miR-16 | RT-qPCR | Upregulated | Breast cancer patients in advanced stages (II, III, and IV) or with distant metastasis have higher expression of miR-155 in serum. | N/A | N/A | N/A | [33] |
| 2 | 2015 | Serum | Four breast cancer patients; four healthy controls | miR-21, miR-34a, miR-125b, miR-155, miR-195, miR-200b, miR-200c, miR-375, and miR-451 | RT-qPCR | No significant different | N/A | N/A | N/A | N/A | [34] |
| 3 | 2016 | Serum | 99 breast cancer patients; 21 healthy controls | miR-21, miR-125b, miR-145, miR-155, and miR-365 | RT-qPCR | Upregulated | miR-155 expression was higher in stage III compared to stage I and II. | 0.749 | 100.00 | 51.02 | [32] |
| 4 | 2016 | Serum | 148 breast cancer patients; 142 heathy controls | miR-155 | RT-qPCR | Upregulated | N/A | 0.879 | 84.20 | 88.10 | [28] |
| 5 | 2016 | Plasma | 106 breast cancer patients; 106 healthy controls | miR-155, miR-21, and miR-10b | RT-qPCR | Upregulated | N/A | 0.692 | 66.00 | 68.90 | [31] |
| 6 | 2017 | Plasma | 128 breast cancer patients; 28 healthy controls | miR-17, miR-18a, miR-19a, miR-20a, miR-21, miR-27a, and miR-155 | RT-qPCR | Downregulated | High expression of miR-155 in invasive breast cancer patients’ plasma was found associated with estrogen and progesterone positive breast cancer. | N/A | N/A | N/A | [35] |
| 7 | 2018 | Plasma | 30 breast cancer patients; 25 healthy controls | miR-21 and miR-155 | RT-qPCR | Upregulated | No correlation between miR-155 and tumor size, lymph nodes, tumor stages and age. | 0.920 | N/A | N/A | [27] |
| 8 | 2018 | Serum | 158 breast cancer patients; 107 healthy controls | Let-7a, miR-155, miR-574-5p, miR-21, miR-10b, miR-181b, miR-1254, miR-196a, miR-205, and miR-195 | RT-qPCR | Upregulated | N/A | 0.817 | 83.30 | 80.00 | [17] |
| 9 | 2019 | Serum | 96 breast cancer patients; 47 benign breast disease; 39 healthy controls | miR-21, miR-155, and miR-126 | RT-qPCR | Upregulated | High expression of miR-155 significantly associated with late stages and high-grade tumor | 0.995 | 95.80 | 96.50 | [30] |
| 10 | 2019 | Plasma | 37 breast cancer patients; 34 healthy controls | miR-155, miR-376c, miR-17a, and miR-10b | RT-qPCR | Upregulated | miR-155 expression was found increased in stage II breast cancer but decreased in stage III and IV breast cancer. However, miR-155 decreases in terms of tumor size, lymph nodes involvement and metastasis in advanced stages. | 0.847 | 100.00 | 73.53 | [25] |
| 11 | 2019 | Serum | 80 breast cancer patients; 40 benign breast disease; 30 healthy controls | miR-17-5p, miR-155, and miR-222 | RT-qPCR | Upregulated | Increased expression of miR-155 in primary BC group. Late stage and advanced grade BC show elevated miR-155 expressions. | 0.933 | 95.00 | 97.10 | [24] |
| 12 | 2020 | Plasma | 102 breast cancer patients; 15 healthy controls | miR-155 | RT-qPCR | Upregulated | miR-155 expression was found higher in patients who are above 40 years old compared to younger group of patients. In addition, high miR-155 expression was observed in patients with grade III breast cancer or tumor size more than 5 cm. | N/A | N/A | N/A | [20] |
| 13 | 2020 | Serum | 36 breast cancer patients; 36 healthy controls | miR-155 | RT-qPCR | Upregulated | Increased expression in breast cancer patients (of all grades, stages, T sizes, lymph node metastasis, PR, ER, HER2, and Ki-67). | 0.890 | 77.78 | 88.89 | [29] |
| 14 | 2020 | Serum | 45 breast cancer patients; 50 idiopathic granulomatous mastitis; 48 healthy controls | miR-155, let-7c, and miR-21 | RT-qPCR | Downregulated | N/A | N/A | N/A | N/A | [36] |
| 15 | 2021 | Plasma | 41 breast cancer patients; 32 healthy controls | miR-155, miR-21, miR-23a, miR-130a, miR-145, miR-425-5p, miR-139-5p, miR-451, miR-195, miR-125b, miR-100, and miR-182 | RT-qPCR | Upregulated | N/A | 0.765 | 78.00 | 75.00 | [21] |
| 16 | 2021 | Plasma | 20 breast cancer patients; 20 healthy controls | miR-21, miR-27b, miR-125a, miR-155, miR-200c, miR-335, and miR-373 | RT-qPCR | Upregulated | N/A | 0.856 | 83.30 | 82.40 | [22] |
| 17 | 2022 | Plasma | 45 breast cancer patients; 15 high risk breast cancer patients; 20 healthy controls | miR-10b, miR-21, miR-155, miR-145, and let-7c | RT-qPCR | Downregulated | miR-155 expression was downregulated in breast cancer patients with lymphatic invasion compared to breast cancer patients without. | 0.836 | 70.00 | 91.00 | [37] |
| 18 | 2022 | Serum | 99 breast cancer patients; 40 healthy controls | miR-155, miR-373, miR-10b, and miR-34a | RT-qPCR | Upregulated | miR-155 was upregulated in PR-negative breast cancer, large tumor and lymph node metastasis. | 0.944 | 86.90 | 90.00 | [23] |
| 19 | 2023 | Serum | 139 triple negative breast cancer patients; 51 healthy controls | miR-205, miR-155, and miR-21 | RT-qPCR | Upregulated | N/A | 0.870 | 87.70 | 63.70 | [26] |
| No. | Year | Sample used | Cohort study | MiRNA | Method | miR-155 expression | Clinicopathological changes | Area under curve of ROC | Sensitivity, % | Specificity, % | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HER2: human epidermal growth factor receptor 2; N/A: not applicable; PR: progesterone receptor; ROC: receiver operating characteristic; RT-qPCR: qualitative real-time polymerase chain reaction. | |||||||||||
| 1 | 2014 | Biopsy | 57 breast cancer patients; 20 healthy controls | miR-148b, miR-10b, miR-21, miR-221, and miR-155 | RT-qPCR | Upregulated | miR-155 was significantly upregulated in PR negative and HER2 overexpressed breast cancer. However, lymph node involvement, tumor size and grade groups did not reveal any significant differences of miR-155 expression. | N/A | N/A | N/A | [39] |
| 2 | 2015 | Urine | 24 breast cancer patients; 24 healthy controls | miR-21, miR-34a, miR-125b, miR-155, miR-195, miR-200b, miR-200c, miR-375, and miR-451 | RT-qPCR | Upregulated | N/A | 0.814 | N/A | N/A | [34] |
| 3 | 2016 | Biopsy | 35 breast cancer patients (adjacent tissue as control) | miR-221, miR-222, miR-20a, miR-200a, miR-155, miR-21, miR-146b, miR-125b, and miR-205 | RT-qPCR | Upregulated | N/A | 0.870 | N/A | N/A | [40] |
| 4 | 2017 | Nipple aspirate fluid | 20 breast cancer patients; 12 healthy controls | miR-16, miR-21, miR-100, miR-129, miR-145, miR-155, miR-181, miR-199, miR-205, and miR-212 | RT-qPCR | No significant different | miR-155 expression was high in node positive breast cancer patients compared to node negative breast cancer patients. | N/A | N/A | N/A | [44] |
| 5 | 2018 | Biopsy | 30 breast cancer patients; 25 healthy controls | miR-21 and miR-155 | RT-qPCR | Upregulated | N/A | 0.810 | N/A | N/A | [27] |
| 6 | 2019 | Biopsy | 50 breast cancer patients; 50 healthy controls | miR-127-3p, miR-133a-3p, miR-155-5p, miR-199b-5p, and miR-342-5p | RT-qPCR | Upregulated | Expression of miR-155 was high in HER-2 positive breast cancer patients. | 0.740 | N/A | N/A | [38] |
| 7 | 2020 | Peripheral blood mononuclear cells | 15 breast cancer patients; 15 healthy controls | miR-155, miR-27b, and miR-29a | RT-qPCR | Upregulated | N/A | 0.941 | 84.40 | 91.10 | [42] |
| 8 | 2021 | Peripheral blood mononuclear cells | 70 breast cancer patients; 40 healthy controls | miR-155 | RT-qPCR | Upregulated | Expression of miR-155 was high with breast cancer with high tumor grade. | N/A | N/A | N/A | [43] |
| 9 | 2022 | Serum exosomes | Seven breast cancer patients; seven healthy controls | miR-21, miR-155, miR-182, miR-373, and miR-126 | RT-qPCR | Upregulated | N/A | N/A | N/A | N/A | [41] |
| Studies | Consecutive or random sample of patient enrolled for control | Avoid inappropriate exclusion? | Were index test results interpreted without knowledge of results of reference standard? | If a threshold was used, was it pre-specified? | Is reference standard likely to correctly classify target condition? | Were the reference standard results interpreted without knowledge of results of index test? | Did all patients receive a reference standard? | Did patients receive same reference standard? | Were all patients included in analysis? | Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 represents “yes”, 0 represents “no” and “unclear”. | ||||||||||
| Nassar et al (2014) [39] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Erbes et al (2015) [34] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Hagrass et al (2015) [33] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Han et al (2016) [32] | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Guo et al (2016) [28] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Zhang et al (2016) [31] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Jurkovicova et al (2017) [35] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Qin et al (2017) [44] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Soleimanpour et al (2018) [27] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Huang et al (2018) [17] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Shaheen et al (2019) [25] | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 7 |
| Kolesnikov et al (2019) [40] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 8 |
| Swellam et al (2019) [24] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Swellam et al (2019) [30] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Aksan et al (2020) [36] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Anwar et al (2020) [20] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Bitaraf et al (2020) [38] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Mojahed et al (2020) [29] | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 6 |
| Raeisi et al (2020) [42] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Itani et al (2021) [21] | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Canatan et al (2021) [22] | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Iranparast et al (2021) [43] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Asgari et al (2022) [41] | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| Mohamed et al (2022) [37] | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 6 |
| Mohamed et al (2022) [23] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
| Kumar et al (2023) [26] | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9 |
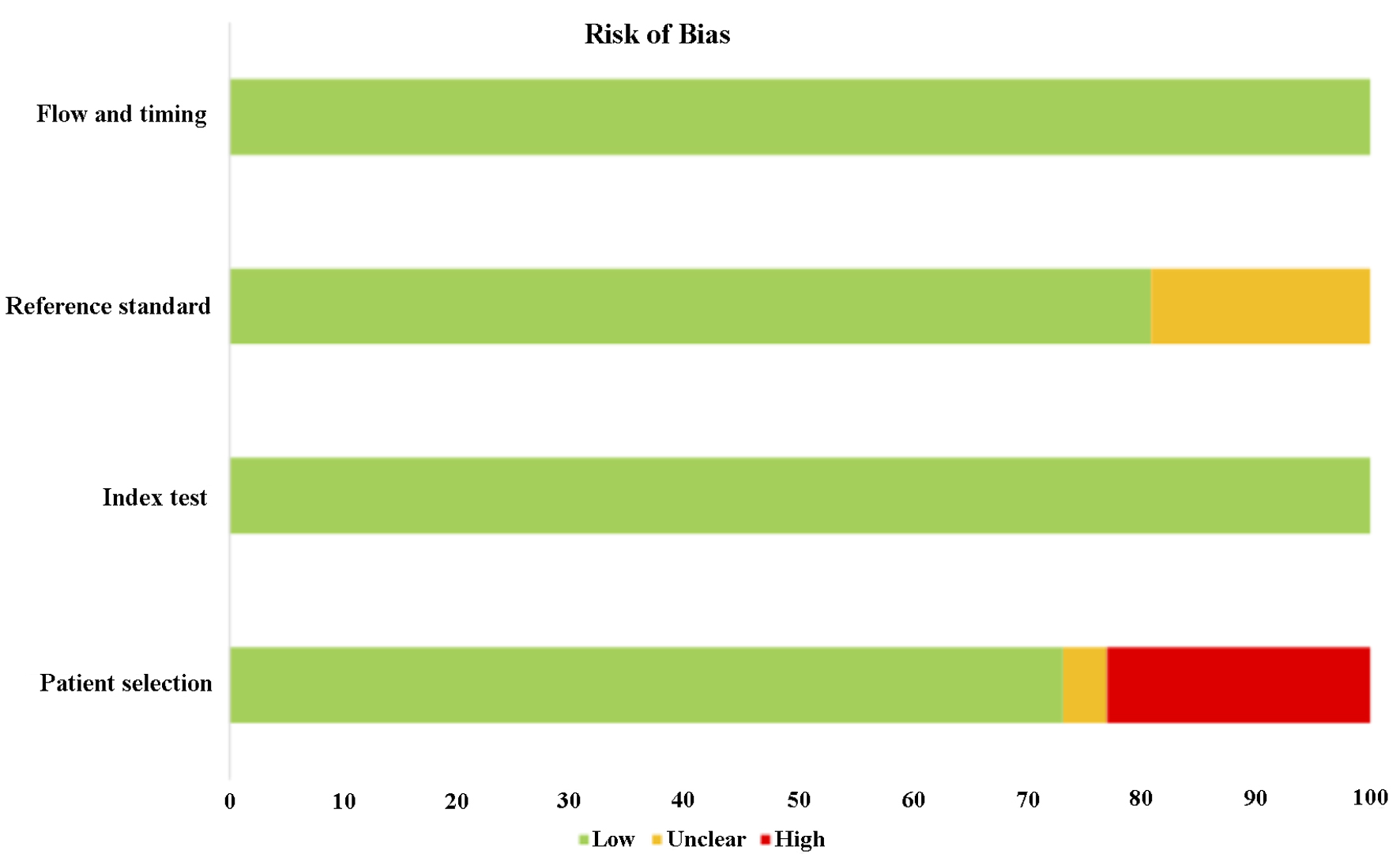
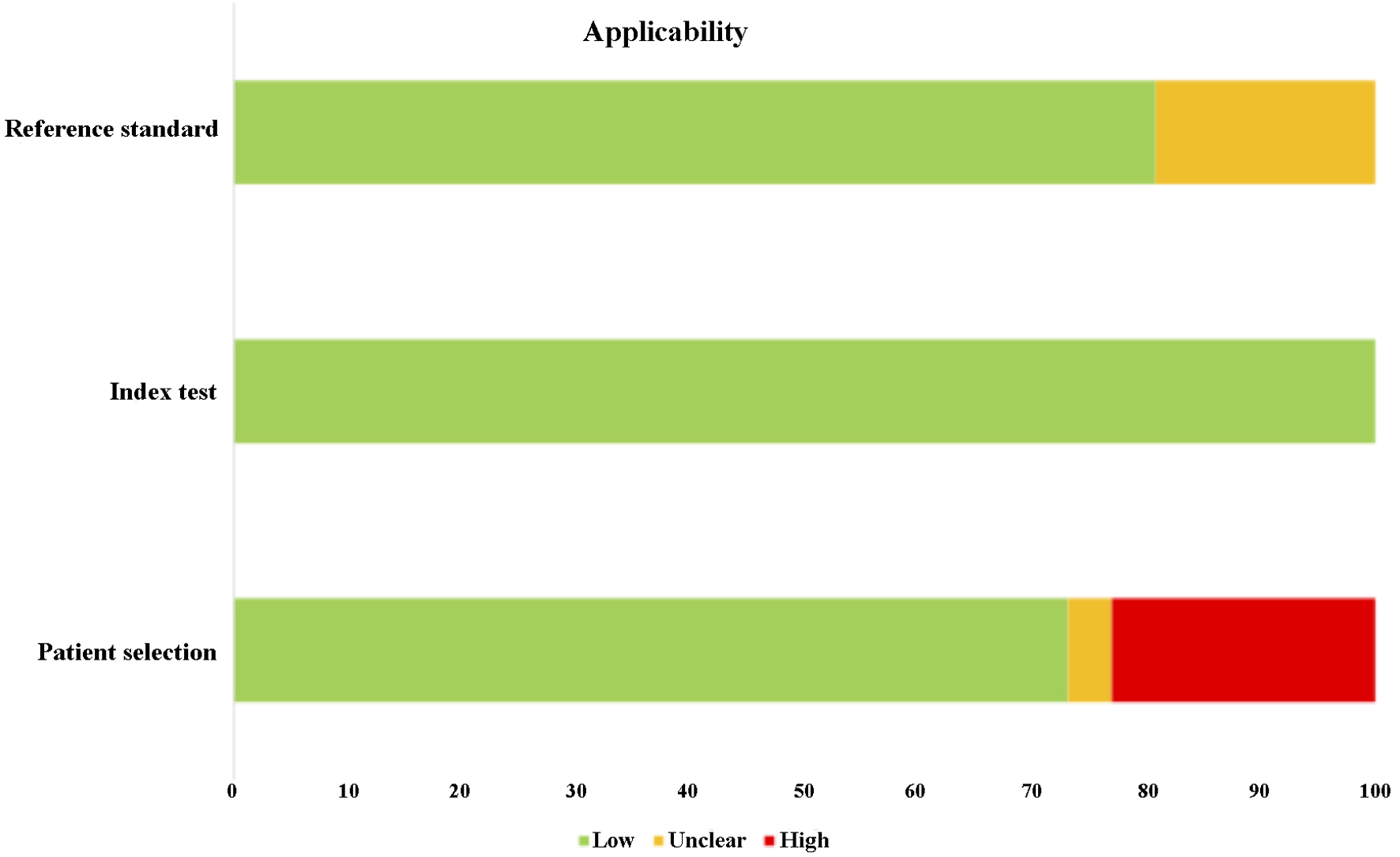
| Risk of bias | Applicability | ||||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| “Low” represents low risk of bias and low risk of errors when methodology is applied; “Unclear” indicates the risk of bias and risk of errors are unclear when the methodology is applied; “High” represents high risk of bias and high risk of errors when methodology is applied. | |||||||
| Nassar et al (2014) [39] | High | Low | Low | Low | High | Low | Low |
| Erbes et al (2015) [34] | Low | Low | Low | Low | Low | Low | Low |
| Hagrass et al (2015) [33] | Low | Low | Low | Low | Low | Low | Low |
| Han et al (2016) [32] | Unclear | Low | Unclear | Low | Unclear | Low | Unclear |
| Guo et al (2016) [28] | Low | Low | Unclear | Low | Low | Low | Unclear |
| Zhang et al (2016) [31] | Low | Low | Low | Low | Low | Low | Low |
| Jurkovicova et al (2017) [35] | Low | Low | Low | Low | Low | Low | Low |
| Qin et al (2017) [44] | Low | Low | Low | Low | Low | Low | Low |
| Soleimanpour et al (2018) [27] | Low | Low | Low | Low | Low | Low | Low |
| Huang et al (2018) [17] | Low | Low | Low | Low | Low | Low | Low |
| Shaheen et al (2019) [25] | High | Low | Low | Low | High | Low | Low |
| Kolesnikov et al (2019) [40] | Low | Low | Low | Low | Low | Low | Low |
| Swellam et al (2019) [24] | Low | Low | Low | Low | Low | Low | Low |
| Swellam et al (2019) [30] | Low | Low | Low | Low | Low | Low | Low |
| Aksan et al (2020) [36] | Low | Low | Low | Low | Low | Low | Low |
| Anwar et al (2020) [20] | Low | Low | Low | Low | Low | Low | Low |
| Bitaraf et al (2020) [38] | Low | Low | Low | Low | Low | Low | Low |
| Mojahed et al (2020) [29] | Low | Low | Unclear | Low | Low | Low | Unclear |
| Raeisi et al (2020) [42] | High | Low | Low | Low | High | Low | Low |
| Itani et al (2021) [21] | High | Low | Low | Low | High | Low | Low |
| Canatan et al (2021) [22] | High | Low | Low | Low | High | Low | Low |
| Iranparast et al (2021) [43] | Low | Low | Low | Low | Low | Low | Low |
| Asgari et al (2022) [41] | High | Low | Unclear | Low | High | Low | Unclear |
| Mohamed et al (2022) [37] | Low | Low | Unclear | Low | Low | Low | Unclear |
| Mohamed et al (2022) [23] | Low | Low | Low | Low | Low | Low | Low |
| Kumar et al (2023) [26] | Low | Low | Low | Low | Low | Low | Low |