| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 6, December 2024, pages 929-941
Interferon Gamma Receptor 2 Collaborates With Circular RNA/MicroRNA to Modulate Programmed Cell Death-Ligand 1 Levels in Nasopharyngeal Carcinoma
Guo Fang Guana, b , Ze Ming Fua, De Jun Zhanga, Ying Yuan Guoa, Fang Guoa, Yi Ning Wana, Jie Baia, Ying Zhaoa
aDepartment of Otolaryngology-Head and Neck Surgery, The Second Hospital of Jilin University, Changchun 130041, China
bCorresponding Author: Guo Fang Guan, Department of Otolaryngology-Head and Neck Surgery, The Second Hospital of Jilin University, Changchun 130041, China
Manuscript submitted October 22, 2024, accepted December 2, 2024, published online December 11, 2024
Short title: Regulation of IFNGR2 Expression in NPC
doi: https://doi.org/10.14740/wjon1994
| Abstract | ▴Top |
Background: The effectiveness of immune checkpoint therapy highlights the need to understand abnormal programmed cell death protein-1 (PD-1) expression in nasopharyngeal carcinoma (NPC), especially when treatments fail, or resistance develops. Interferon gamma (IFN-γ) signaling is crucial for regulating programmed cell death-ligand 1 (PD-L1) expression. Our study focuses on interferon gamma receptor 2 (IFNGR2), an essential part of the IFN-γ pathway, and its impact on malignant traits in NPC.
Methods: The expression levels of IFNGR2 and PD-L1 were accessed using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). To understand the cellular phenotypic effects, small interfering RNA (siRNA)/short hairpin RNA (shRNA) knockdown techniques were used to evaluate cell viability, clonogenic survival, migration and invasion, immunohistochemistry, and tumor formation assays. The relationship between IFNGR2 and microRNAs (miRNAs)/circular RNAs (circRNAs) will be verified using methods such as circRNA stability assay, rescue, and dual-luciferase reporter assay.
Results: IFNGR2 was significantly overexpressed in NPC, and its expression positively correlated with PD-L1 levels. This overexpression contributed to increased cell proliferation, migration, invasion, clonogenicity, and tumor growth. Additionally, we identified an oncogenic circular RNA, circ_001377, and uncovered a novel mechanism by which upregulation of circ_001377 competitively bound to miR-498-3p. This interaction reduced miR-498-3p’s ability to target IFNGR2. As a result, the diminished miR-498-3p led to increased IFNGR2 expression, which subsequently activated the IFN-γ signaling pathway and drove abnormal PD-L1 expression.
Conclusions: IFNGR2 is an oncogenic factor in NPC. The circ_001377/miR-498-3p interaction drives IFNGR2 upregulation and PD-L1 overexpression, suggesting that targeting this axis could improve therapeutic outcomes.
Keywords: IFNGR2; PD-L1; microRNA; circRNA; Nasopharyngeal carcinoma
| Introduction | ▴Top |
Nasopharyngeal carcinoma (NPC), a distinct non-keratinizing cancer affecting the head and neck, is notably linked to Epstein-Barr virus (EBV) infection in almost all cases [1, 2]. This association with EBV, along with various genetic factors, complicates the treatment and research of NPC [3]. Despite advancements in multimodal therapies, such as surgery and intensity-modulated radiotherapy, which have significantly enhanced local disease management, about 30-40% of patients with more aggressive forms of the disease experience distant metastases within 5 years, often due to chemoradiotherapy resistance [4, 5]. In recent developments, programmed cell death protein-1 (PD-1)/programmed cell death-ligand 1 (PD-L1) checkpoint inhibitor immunotherapy has shown promising outcomes in boosting anti-tumor immune responses in certain cancers. Yet only a minority of head and neck cancer patients, including those with NPC, achieve remission through these treatments [6]. With high recurrence and metastasis rates in NPC, there is a pressing need to investigate the mechanisms underlying immunotherapy resistance. Understanding these could pave the way for novel therapeutic strategies in NPC management.
The PD-1/PD-L1 pathway represents one of the key strategies by which EBV-associated NPC evades the immune system [6]. Current research highlights that PD-L1 expression is primarily triggered by interferon gamma (IFN-γ), an essential cytokine in the immune response. IFN-γ interacts with the interferon gamma receptors 1 and 2 (IFNGR1/2), activating the JAK1 and JAK2 enzymes. Concurrently, IFN-γ aids in the recruitment and nuclear translocation of STAT proteins, which, along with JAK1 and JAK2, stimulate PD-L1 expression [7]. Notably, IFNGR2 emerges as a pivotal player in this process, being crucial for recognizing IFN-γ at the forefront, thereby enhancing the body’s ability to defend against infections and cancers. Elevated levels of IFNGR2 have been identified in conditions such as glioblastoma [8], where its upregulation is linked to increased cell proliferation, invasion, and migration in glioma models in vitro [9]. Similarly, in ovarian cancer, the reduction of IFNGR2 led to a decrease in PD-L1 expression, underscoring the dependency of PD-L1 induction on IFN-γ receptors [10]. Additionally, IFNGR2 has been characterized as a novel suppressor of the pro-apoptotic protein Bax, showcasing anti-apoptotic properties [11]. Despite these insights, the full scope of how IFN-γ signaling genes contribute to immune responses against tumors, especially in the context of resistance to PD-1/PD-L1 checkpoint blockade therapy, remains an area ripe for further exploration.
MicroRNAs (miRNAs) and circular RNAs (circRNAs) are two classes of non-coding RNAs that play essential roles in gene expression regulation. There is considerable evidence highlighting the involvement of various miRNAs in the regulation of NPC. Notably, EBV-encoded miRNAs, such as the overexpressed BART miRNAs, have gained attention for their potential as noninvasive biomarkers in the diagnosis and prognosis of NPC [12]. Studies on miRNA expression profiles in NPC have unveiled a 5-miRNA signature (comprising miR-93, miR-142-3p, miR-29c, miR-26a, and miR-30e) that correlates with overall survival (OS) and disease-free survival (DFS) [13]. Similarly, circRNAs are increasingly recognized for their profound influence on cancer biology, including roles in diagnosis, prognosis, and treatment. Their ability to act as miRNA sponges, effectively sequestering miRNAs and influencing gene expression, underscores their significance in cellular functioning [14]. For example, circ_0003738 has been shown to modulate IFNGR2 expression in psoriasis by sponging miR-490-5p [15]. Furthermore, miR-142-3p can reduce IFNGR2 expression by directly targeting evolutionarily conserved sequences within their 3′-untranslated regions (UTRs) [16]. Despite these insights, the specific roles and mechanisms of action of most miRNAs and circRNAs in regulating IFNGR2 expression in NPC are still largely unexplored. This gap in knowledge presents a rich area for future research, potentially uncovering new therapeutic targets and diagnostic markers. In this study, we propose a molecular pathway in which the aberrant expression of circ_001377 acts as a sponge for miR-498-3p. This interaction attenuates IFNGR2 repression, leading to increased IFNGR2 expression, which in turn leads to increased PD-L1 levels in NPC through the IFN-γ pathway. The study aims to enhance the understanding of the pathogenesis of NPC and identify new potential therapeutic avenues for its treatment.
| Materials and Methods | ▴Top |
Cell lines
Two STR genotyping verified human NPC cell lines were purchased from Shanghai Yubo Biotech Co., China. The EBV-negative NPC cell line HK1 and EBV-positive cell line C666-1 were used in the study. These were cultured in RPMI medium enriched with 10% fetal bovine serum (FBS). Additionally, the non-tumorigenic human nasopharyngeal cell line NP69, which has been immortalized with SV40, was cultured in keratinocyte-free media with added epidermal growth factor (EGF) and pituitary serum (Thermo Fisher Scientific, Shanghai, China) serving as the normal control. All cells were maintained in a 37 °C incubator with 5% of CO2 and humidify and were confirmed to be free from mycoplasma contamination.
RNA extraction and quantification of mRNAs, miRNAs and circRNAs
RNA was isolated from cell lines using the Qiagen RNA extraction kit according to the manufacturer’s instructions. The total RNA was reverse transcribed by the SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time polymerase chain reaction (PCR) analysis was performed using SYBR Green PCR Master Mix (Thermo Fisher Scientific, Inc.) and the ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The expression of miRNAs and circRNAs was detected as previously described [17]. RNU44 and RNU48 were used as endogenous controls. The TaqMan MicroRNA Assay included a reverse transcription step in which a stem-loop reverse transcription primer specifically hybridized to the miR-498-3p molecule, and a mimic of miR-498-3p were purchased from Thermo Fisher Scientific. The primers used in the study are as follow: IFNGR2: forward 5′-CTCCATTCTGCCTGGGTGACAA-3′, reverse 5′-CGTGGAGGTATCAGCGATGTCA-3′; PD-L1: forward 5′-TGCCGACTACAAGCGAATTACTG-3′, reverse 5′-CTGCTTGTCCAGATGACTTCGG-3′; Circ_001377: forward 5′- CTGGGGATTCCAAGATCAGA-3′, reverse 5′-ATAGCGTTTGTGGGCTTTGT-3′.
Cell transfection, proliferation, and clonogenic assays
The LipofectAMINE RNAiMAX (Life Technologies) reverse transfection protocol was used to perform RNA interference (RNAi) experiments, according to the manufacturer’s instructions. IFNGR2-target siRNA, premiR-498-3p, siRNA for circ_001377, and the negative control (NC) siRNA were purchased from Thermo Fisher Scientific (Shanghai, China). The pLV-circ vector carrying the overexpressing circ_001377 plasmids were designed and synthesized by GenePharma (Shanghai, China).
NPC cells were seeded into 96-well plates for cell proliferation assays or six-well plates for Western blot and transfected with the reagents above. The cytotoxicity effects of HK1 and C666-1 cells post-transfection were evaluated using the CyQUANT® NF Cell Proliferation Assay Kit (C35006, Invitrogen). Cell proliferative activity was measured at 24, 48 and 72 h after transfection.
NPC cells were transfected into 12-well plates for colony formation assay. After 48 hs, cells were harvested, and 300 or 500 cells were re-seeded onto six-well plates in triplicate. After a 2-week culture, the plates were fixed and stained, and the number of colonies was counted using a microscopy. The fraction of viable cells was calculated by comparison with transfected cells or with scramble controls (SC).
Cell migration and invasion assays
To investigate the cellular effects of knockdown IFNGR2, HK1 and C666-1 cells were transfected with siRNA target to IFNGR2 or siRNA SC and plated on either the six-well control inserts (PET membrane) or trans-well invasion chambers pre-coated with Matrigel (BD Biosciences). A medium containing 15% FBS in the lower chamber served as the chemo-attractant. The plates were incubated for 24 h. After removing non-migrating or invading cells from the upper surface of the membrane with cotton swabs, the migrating or invasive cells attached to the low surface of the membrane insert were then fixed and stained with Diff-Quick Stain (BD Biosciences). A light microscope was used to count migrating or invasive cells.
Immunohistochemistry for Ki-67 and CD31
The expressions of Ki-67 and CD31 were evaluated on xenograft tumor sections. Mouse anti-human Ki-67 antigen (1:100 dilution; DakoCytomation) and rat anti-mouse CD31 antibody (1:50 dilution; BD Pharmingen) were utilized. A microwave antigen retrieval method in combination with the Level-2 Ultra Streptavidin system (DakoCytomation) was used. Ki-67 positive nuclei staining was evaluated in the regions with the highest expression, referred to as tumor hot spots. Three hot spots were selected at random, and at least 100 tumor cells were counted in each. The final score was determined by calculating the average percentage of Ki-67 positive cells across the three hot spots. For CD31 evaluation, as described previously [18], using low-power (× 100) microscopy, the three areas with the highest vascularization were selected. Vessels with a clearly defined lumen or distinct linear shape were counted. The final score was calculated as the average vessel count from these three fields.
Western blotting
Cells from various transfections were harvested and lysed 72 h after transfection. Protein concentrations were determined using the bicinchoninic acid (BCA) assay. Each sample, containing 20 µg of protein, was separated on 10% Tris-glycine SDS-PAGE gels and subsequently transferred to nitrocellulose membranes. The membranes were then blocked using 5% milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) before being incubated overnight with primary antibodies: rabbit anti-IFNGR2 (PA5-141049, 1:500, Thermo Fisher-Invitrogen) and mouse anti-beta Actin (ab8226, Abcam, USA). This step was followed by a 2-h incubation with horseradish peroxidase-conjugated secondary antibodies (Abcam, USA). Protein bands were detected using the ECL Western blotting detection system (Pierce, #34095).
Luciferase reporter assay
The Dual-report luciferase assays system was used to investigate the interactions between miRNAs and IFNGR2, as well as between circRNAs and miRNAs. PCR amplified the sequences of both the wild-type and mutant versions of the IFNGR2 3′-UTR regions, which include the presumptive binding sites for miR-498-3p. In a similar manner, sequences of the wild-type and mutant circ_001377 that contain miR-498-3p binding sites were amplified. These amplified sequences were then cloned into pmiRreport luciferase vectors (Ambion). Subsequently, cells were co-transfected with these vectors along with miR-498-3p mimics or SC. This setup facilitated the detection of direct interactions between the 3′-UTR of IFNGR2 or circ_001377 and miR-498-3p through the co-transfection of miRNA mimics in C666-1 cells. For normalization purposes, each well was also transfected with the pRL-SV40 vector (Promega), which contains Renilla luciferase. Luciferase activity was measured 48 h post-transfection using the Dual-Glo Luciferase Assay System (Promega).
RNase R treatment and actinomycin D assay
Total RNA (2 µg) from C666-1 cells was treated with or without 3 U/µg RNase R (Geneseed Biotech Co., Ltd., Guangzhou, China) to eliminate genomic DNA. The levels of linear and circular RNA were then determined via quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). To evaluate the stability of circ_001377, C666-1 cells were treated with actinomycin D (100 ng/mL) (Merck, Germany) at various time points: 0 h, 4 h, 8 h, 12 h, and 24 h. Subsequent to these treatments, total RNA was extracted and the expression levels of circ_001377 and MRPS15 mRNA were quantified using qRT-PCR.
Animal experiments
Short hairpin RNA (shRNA) targeting IFNGR2 and a non-targeting control shRNA (NC) were synthesized by GenePharma (Shanghai, China). These shRNA constructs were introduced into HEK-293T cells using co-transfection with pGPU6/Hygro and pGPH1 plasmids. The lentiviral particles produced were collected from the cell culture supernatant and used to infect C666-1 cells, which were then selected with puromycin (2 µg/mL). Severe combined immunodeficiency (SCID) mice were divided into two groups: the NC group and the shIFNGR2 group, with five mice in each group. Tumor growth was monitored bi-weekly, following previously described procedures [17]. Mice were euthanized when tumors reached a predetermined size. The volume of the tumors was calculated using the formula: volume = (width2 × length)/2. All procedures involving animals were conducted in accordance with the guidelines set by the Institutional Animal Care and Use Committee (IACUC) of Jilin University (approval number: SY202210015).
Statistical analysis
Data are presented as mean ± standard error (SE). Statistical analyses were conducted using GraphPad Prism version 9.0 (GraphPad Software, CA, USA). Differences between groups were assessed using Student’s t-test. P values were two-tailed, with a significance level set at α = 0.05. Results represent findings from three independent experiments.
| Results | ▴Top |
IFNGR2 expression was elevated in NPC and contributed to enhanced cell proliferation and clonogenic potential
The expression level of IFNGR2 was assessed by qRT-PCR in NPC cells. Results revealed a significantly upregulation of IFNGR2 in both HK1 and C666-1 cells compared to the control group (P < 0.05) (Fig. 1a). Given the pivotal role of the IFNGR1/2 in mediating the IFN-γ-JAK-STAT pathway and subsequent PD-L1 expression, PD-L1 levels were also examined in these NPC cells. As anticipated, PD-L1 exhibited pronounced expression in both NPC cell lines (P < 0.05) (Fig. 1b). Further exploration into the pathological impact of IFNGR2 was conducted using siRNA-mediated knockdown, which significantly diminished cell viability in both NPC cell models at 48 h post-transfection (P < 0.05) (Fig. 1c, d), as measured by the CyQUANT® NF cell proliferation assay. The functional relevance of IFNGR2 was also evidenced through colony formation assays, where siRNA against IFNGR2 notably decreased the number of colonies compared to the control or scramble siRNA-treated groups (P < 0.05) (Fig. 1e, f).
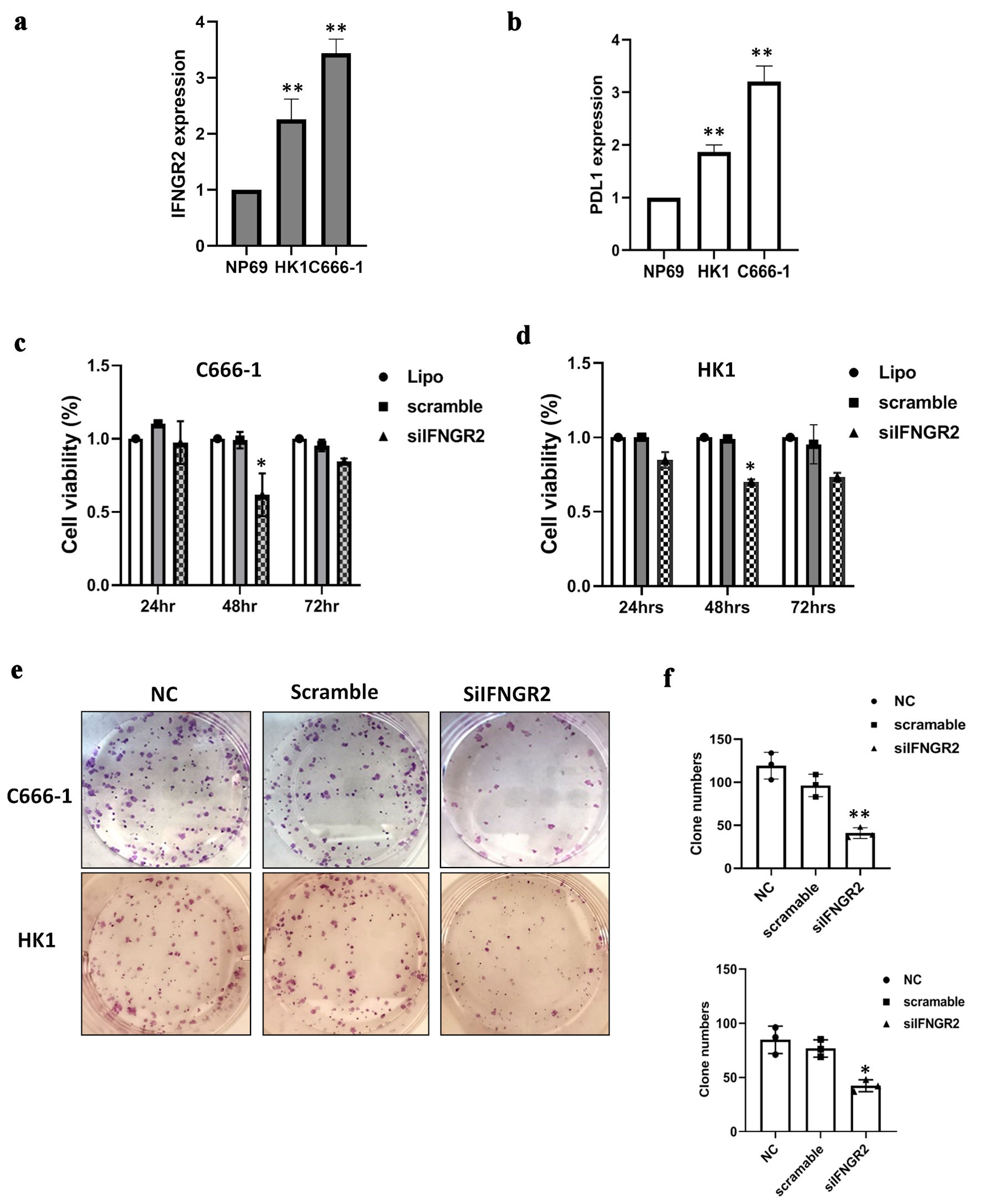 Click for large image | Figure 1. IFNGR2 was upregulated in NPC and promoted cell proliferation and clonogenicity. (a) IFNGR2 overexpression in two NPC cell lines. (b) An increase in PD-L1 levels was noted in NPCs. (c, d) SiRNA targeting IFNGR2 was introduced into HK1 and C666-1 cells, with cell viability assessed at 24, 48, and 72 h post-transfection using the CyQUANT® NF Cell Proliferation Assay. (e, f) Depletion of IFNGR2 significantly reduced the capacity for colony formation. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; NPC: nasopharyngeal carcinoma; PD-L1: programmed cell death-ligand 1; NC: negative control; siRNA: small interfering RNA. |
Effects of IFNGR2 silencing on cell migration, invasion and in vivo tumor formation
Further investigations were conducted to elucidate the effects of IFNGR2 suppression on the cytotoxic properties of transfected cells. The influence of IFNGR2 on the migratory and invasive capabilities of NPC cells was evaluated using Transwell migration assays and Matrigel invasion assays. The reduction of IFNGR2 led to a significant decrease in both cell migration and invasion, with a mean reduction of 55% in migration and 46% in invasion observed in both HK1 and C666-1 cells (P < 0.05) (Fig. 2a, b). To explore the role of IFNGR2 in the formation of NPC tumors in a live model, C666-1 cells with stable IFNGR2 suppression were subcutaneously injected into SCID mice. The progression and volume of the tumors were monitored, revealing that the depletion of IFNGR2 decelerated tumor growth when compared to the control group (P < 0.05) (Fig. 2c, d). Moreover, immunohistochemical analysis for Ki-67 and CD31 expression in the xenograft tumor models showed lower expression levels of both markers in the IFNGR2-suppressed group than in the control group (P < 0.05) (Fig. 2e, f). These results suggest that silencing IFNGR2 impedes tumor development and diminishes the risk of metastasis.
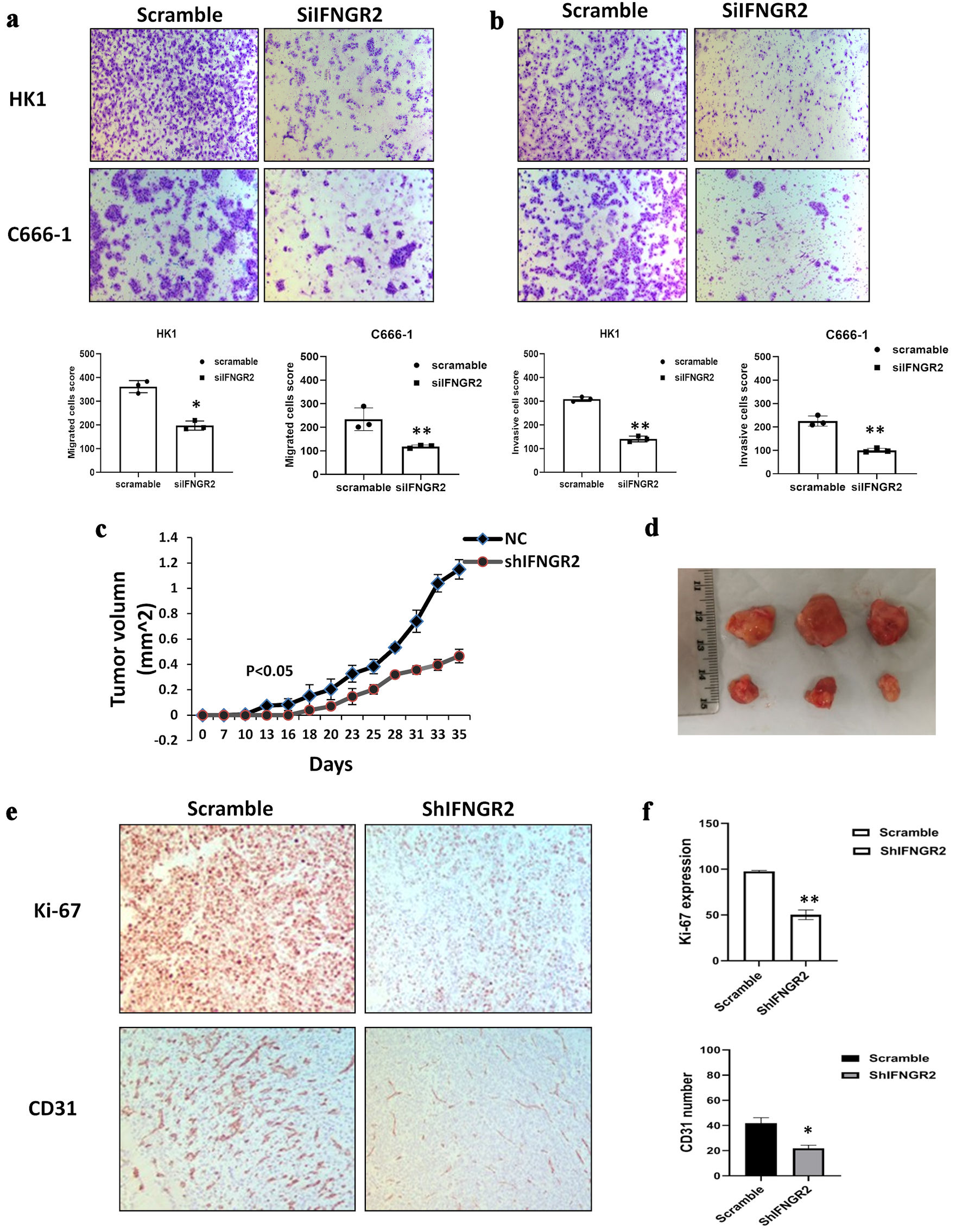 Click for large image | Figure 2. IFNGR2 knockdown inhibited cell migration, invasion and reduced tumor growth in vivo. (a, b) Representative images and quantification of the reduction in migratory capacity (left panel) and invasion (right panel) of HK1 and C666-1 cells that were transfected with IFNGR2-targeting siRNA, compared to negative SC. (c, d) Depletion of IFNGR2 delayed tumor growth compared with the control. Tumor volume was measured. (e, f) Immunohistochemistry detected Ki-67 and CD31 expression in IFNGR2 knockdown group compared with the controls. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; NC: negative control; siRNA: small interfering RNA; SC: scramble control. |
IFNGR2 was a directly target of miR-498-3p
To better understand the molecular intricacies of IFNGR2, we employed several gene target prediction platforms (miRWalk, miRDB, miRSystem, miRSearch and MicroT-CDS), integrating them with insights into their diverse roles in tumor dynamics and cellular mechanisms. This comprehensive approach led to the identification of five miRNAs with potential tumor-suppressing functions, which might be implicated in the regulation of IFNGR2. Initial qRT-PCR analyses were conducted to gauge the expression levels of these miRNAs within the cells. Results depicted in Figure 3a revealed that, of the miRNAs tested, miR-498-3p and miR-497-5p exhibited a pronounced reduction in expression in both HK1 and C666-1 cells in comparison to NP69 (normal control) cells. This pattern suggests a regulatory association between the upregulation of IFNGR2 and the activity of miR-498-3p or miR-497-5p. Figure 3b highlighted the shared binding sites on the 3′-UTR of IFNGR2 for hsa-miR-498. To investigate the direct engagement between the miRNAs and IFNGR2, we engineered various luciferase reporter vectors incorporating the suspected binding sites downstream of the firefly luciferase reporter gene within the pMIR-report vector, as previously outlined [19]. Following this, C666-1 cells were co-transfected with either premiR-498-3p or premiR-497-5p alongside either the pmiR-IFNGR2 3′-UTR or its mutated counterpart. Compared to the controls, the dual luciferase reporter assay showed that overexpression of miR-498-3p significantly reduced luciferase activity in cells transfected with wide-type IFNGR2 3′-UTR, while the mutant construct showed no significant change (Fig. 3c). However, the cells transfected with miR-497-5p did not affect luciferase activity. These findings from the dual-luciferase reporter assays substantiate a direct interaction between miR-498-3p and the 3′-UTR of IFNGR2.
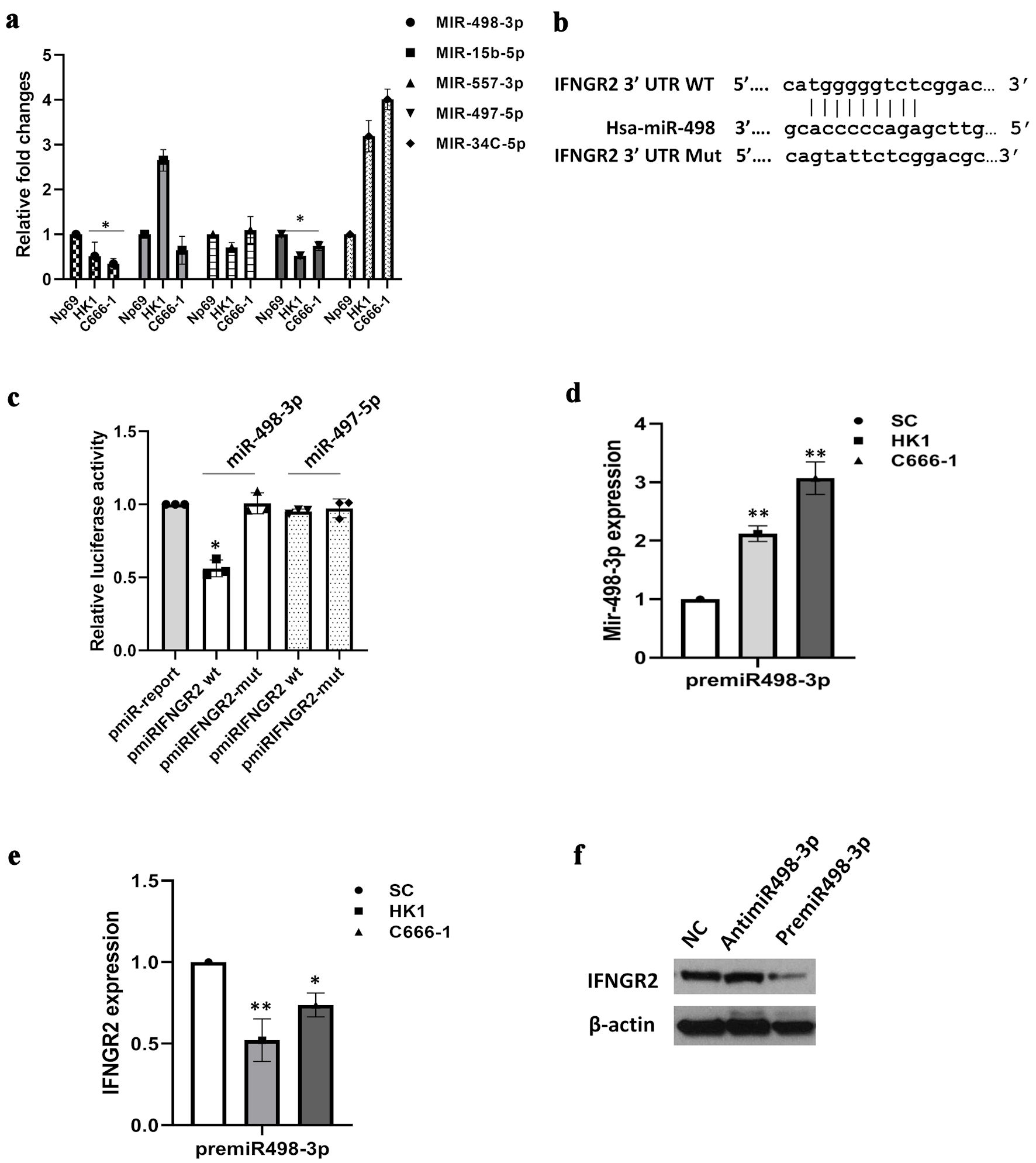 Click for large image | Figure 3. IFNGR2 was directly targeted by miR-498-3p. (a) Bioinformatics analyses led to the identification of five potential miRNA regulators of IFNGR2, with qRT-PCR validations conducted subsequently. (b) Diagram illustrating the binding regions of miR-498-3p on the IFNGR2 sequence, highlighting both wild-type (WT) and mutated (mut) configurations. (c) A dual-luciferase assay verified the specific interaction between miR-498-3p and IFNGR2. (d) Introduction of miR-498-3p mimics elevated miR-498-3p levels in HK1 and C666-1 cells 48 h after transfection. (e) Transfecting cells with premiR-498-3p led to a decrease in IFNGR2 levels. (f) The influence of miR-498-3p on IFNGR2 expression was further confirmed through Western blot analysis. *P < 0.05; **P < 0.01. SC: scramble control; IFNGR2: interferon gamma receptor 2; UTR: untranslated region; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction. |
The interaction between miR-498-3p and IFNGR2 was explored in greater depth. Introducing premiR-498-3p into NPC cells significantly elevated miR-498-3p levels in both HK1 and C666-1 cells, while concurrently diminishing IFNGR2 expression (P < 0.05) (Fig. 3d, e). Western blot analysis further corroborated these findings, demonstrating that while the use of antimiR-498-3p left IFNGR2 expression unaltered, the introduction of premiR-498-3p resulted in a notable decrease in IFNGR2 protein levels (Fig. 3f).
Circ_001377 functioned as a sponge for miR-498-3p
Numerous studies have indicated that circRNAs may indirectly influence gene expression by acting as miRNA sponges. In exploring the potential relationship between circRNAs and miR-498-3p, we employed bioinformatics tools (TargetscanHuman 7.2 and the circular RNA interactome) and analyzed a microarray dataset (GSE190271 from the Gene Expression Omnibus). We identified the top five upregulated circRNAs based on the number of miR-498-3p binding sites and their context scores. Subsequent KEGG pathway analysis led us to select hsa_circ_001377 (also known as hsa_circ_0000054, gene symbol: MRPS15) as a circRNA of interest for further investigation (Fig. 4a). The identified binding sites between circ_001377 and miR-498-3p were depicted in Figure 4b. Initially, we assessed the stability of circ_001377 in NPC cells using RNase R treatment. The findings revealed that circ_001377 exhibited notable resistance to RNase R degradation, whereas the levels of linear RNA (GAPDH) were significantly diminished (Fig. 4c). Additionally, when cells were treated with the transcription inhibitor actinomycin D, circ_001377 showed remarkable stability compared to the linear mRNA (MRPS15) (Fig. 4d). We also measured the expression levels of circ_001377 in NPC cell lines using qRT-PCR. The data indicated that the expression levels of circ_001377 in HK1 and C666-1 cells were significantly elevated compared to those in normal control samples (Fig. 4e). Furthermore, we evaluated the binding interaction between miR-498-3p and circ_001377 using a dual-luciferase reporter assay. Constructs of both wild-type and mutant versions of the circ_001377 luciferase reporter system were developed, and miR-498-3p mimics or scramble miRNA were transfected into C666-1 cells. The findings demonstrated that the introduction of miR-498-3p markedly decreased luciferase activity in the construct containing wild-type circ_001377, whereas no significant effect was observed on the mutant construct (Fig. 4f). Collectively, these results indicate that circ_001377 plays a crucial role in NPC by modulating the activity of miR-498-3p.
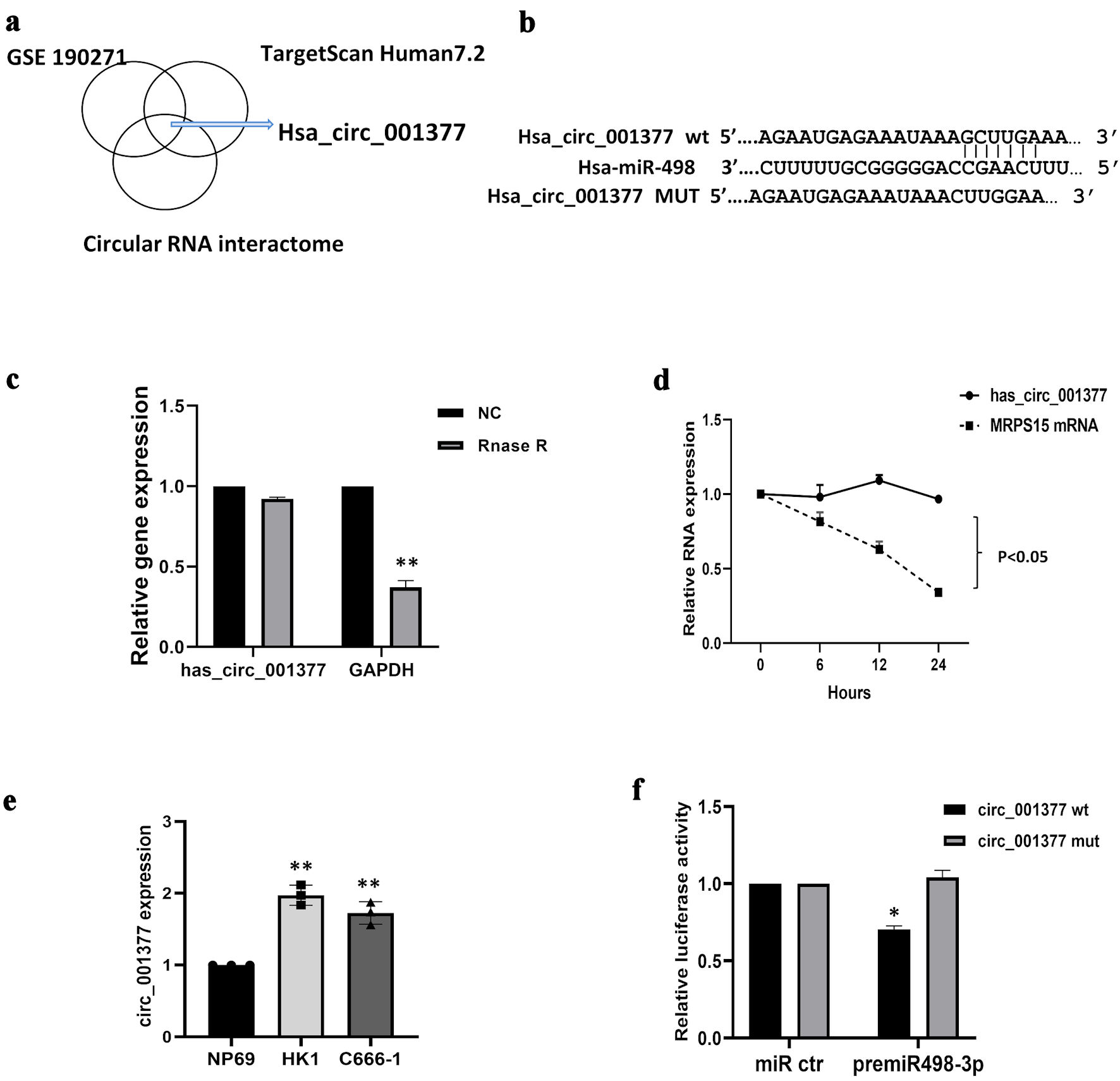 Click for large image | Figure 4. Circ_001377 acted as a sponge for miR-498-3p. (a) Circ_001377 identification involved circular RNA interaction predictors, TargetScan Human7.2, and GSE190271 dataset analysis, complemented by KEGG pathway analysis. (b) Diagram depicting the miR-498-3p binding site on the wild-type (WT) circ_001377 sequence alongside a mutant variant of circ_001377. (c) Following RNase R or mock treatment of C666-1 cell-derived RNA, circ_001377 stability was assessed via qRT-PCR. (d) C666-1 cells were treated with actinomycin D, the relative mRNA levels were measured by qRT-PCR. (e) Circ_001377 expression was measured across NPC cell lines. (f) Luciferase assay outcomes in C666-1 cells transfected with luciferase reporters linked to miR-498-3p targets on WT or mutated circ_001377 sites, alongside miR-498-3p mimics or controls. *P < 0.05; **P < 0.01. NC: negative control; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction; NPC: nasopharyngeal carcinoma. |
Circ_001377 collaborating miR-498-3p and IFNGR2 regulated PD-L1 expression
Further exploration into the interplay between miR-498-3p and circ_001377 regarding their influence on IFNGR2 revealed intricate regulatory mechanisms. Transfection experiments using overexpressed circ_001377 constructs, premiR-498-3p, or a combination thereof were conducted in HK1 and C666-1 cells. The qRT-PCR analysis indicated a significant elevation in miR-498-3p levels upon the introduction of its mimic, whereas circ_001377 overexpression led to a reduction in miR-498-3p levels. However, this effect was mitigated when both premiR-498-3p and circ_001377 constructs were co-transfected, as demonstrated by a recovery in miR-498-3p levels compared to the NC (P < 0.05) (Fig. 5a). Similarly, IFNGR2 levels were notably upregulated following circ_001377 plasmid transfection but were decreased upon premiR-498-3p introduction. This increase in IFNGR2 expression was subsequently moderated by the combined transfection of premiR-498-3p and circ_001377 plasmids (P < 0.05) (Fig. 5b). Additionally, experiments involving the introduction of premiR-498-3p, si-circ-001377, or both showcased a significant reduction in IFNGR2 expression in cells with premiR-498-3p and circ_001377 knocked down, compared to the NC. This downregulation of IFNGR2 was counteracted upon co-transfection with both premiR-498-3p and si-circ-001377, as evidenced by the Western blot analysis (Fig. 5c). In corresponding studies, PD-L1 expression mirrored the trend observed with IFNGR2. Notably, PD-L1 expression increased following circ_001377 plasmid transfection but diminished upon premiR-498-3p introduction (Fig. 5d). These data reveal a new regulatory mechanism, in which circ_001377 may increase IFNGR2 levels by acting as a sponge for miR-498-3p, thereby indirectly affecting PD-L1 expression in NPC cells, as illustrated in Figure 5e.
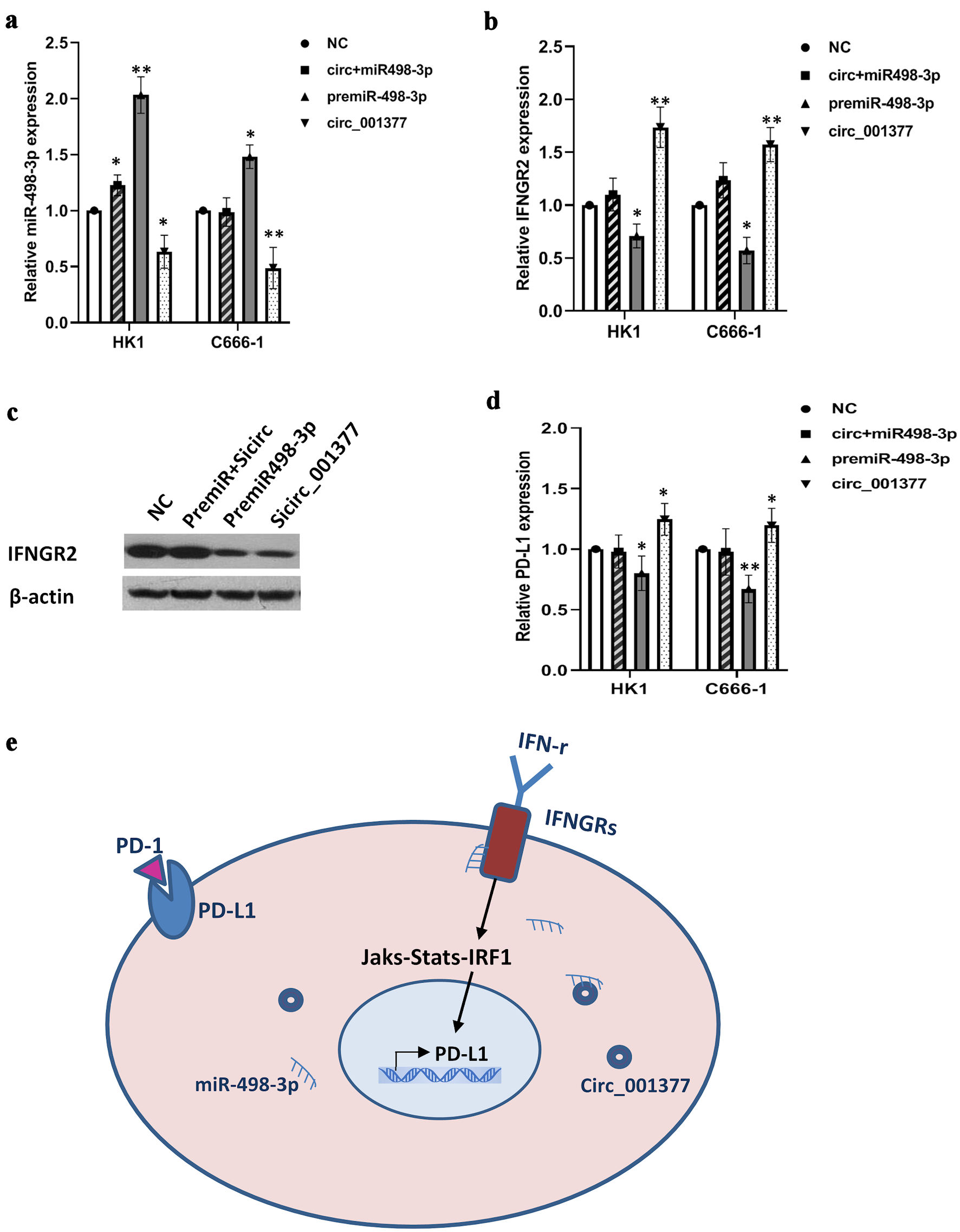 Click for large image | Figure 5. Circ_001377 bound to miR-498-3p, reducing its interaction with IFNGR2 and regulating PD-L1 expression. (a, b) Rescue experiments conducted with premiR-498-3p, circ_001377 overexpression plasmids, or a combination thereof, followed by qRT-PCR assessment of miR-498-3p and IFNGR2. (c) Alterations in IFNGR2 expression caused by miR-498-3p enhancement or circ_001377 depletion were assessed by Western blotting. (d) PD-L1 expression was influenced by the introduction of premiR-498-3p, overexpression plasmids for circ_001377, or a combination of both. (e) Schematic of a proposed model where circ_001377 functions as a sponge to sequester miR-498-3p, resulting in increased IFNGR2 levels. This, in turn, enhanced the IFN-γ signaling pathway, leading to the overexpression of PD-L1 in NPC. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction; NC: negative control; PD-L1: programmed cell death-ligand 1; IFN-γ: interferon gamma; PD-1: programmed cell death protein-1. |
| Discussion | ▴Top |
PD-1/PD-L1 immune checkpoint blockade therapy has achieved remarkable success in inducing tumor regression. Yet, secondary resistance often emerges as immune-selective pressure fosters the development of drug-resistant tumor clones, resulting in a subset of head and neck squamous cell carcinoma (HNSCC) patients, including those with NPC, experiencing minimal treatment response [4, 20]. Consequently, it is imperative to elucidate the mechanisms by which key genes in the IFN-γ signaling pathway contribute to the abnormal expression of PD-L1 and to adjust treatment strategies accordingly. In this study, we revealed that IFNGR2, a significant cytokine receptor, is highly expressed in NPC cells. The upregulation of IFNGR2 was found to enhance tumor cell proliferation and colony formation abilities in HK1 and C666-1 cell lines. Conversely, silencing IFNGR2 attenuated the migratory and invasive capabilities of the tumor cells, reduced tumor growth, and inhibited tumor cell proliferation and angiogenesis in vivo.
Mounting evidence indicates that PD-L1 is frequently overexpressed in various human cancers, yet the precise mechanisms driving its overexpression remain largely unexplored. IFN-γ is recognized as a critical cytokine produced by T cells, playing a pivotal role in immune regulation. Recent research has pinpointed disruptions in the IFN-γ signaling pathway in tumor cells resistant to drugs among patients, who have shown disease progression following anti-PD-1 therapy [6, 21]. This phenomenon is particularly intriguing given the dual role of IFN-γ in modulating anti-tumor immunity, both promoting and inhibiting immune responses. IFN-γ triggers the upregulation of essential negative regulatory molecules by engaging the IFNGR1/2, activating JAK1 and JAK2, and subsequently mobilizing STAT to regulate PD-L1 expression [22]. Additionally, IFN-γ contributes to T-cell apoptosis during in vivo immune responses [22]. The human IFNGR2 gene is situated on chromosome 21, and studies on mouse gene promoters indicate that IFNGR2 possesses numerous sites for transcription factor activation, suggesting potential regulation of IFNGR2 expression [23]. IFNGR2 plays a role in managing various immunosuppressive conditions. For instance, single-cell RNA sequencing has unveiled that dendritic cell migration in human sepsis is triggered through TNFRSF-NF-κB and IFNGR2-JAK-STAT3-dependent pathways [24]. Notably, evidence suggests that genetic abnormalities in IFNGR2 may contribute to the risk of cancer development in human colon and rectal cancers [25]. Furthermore, high expression levels of RUNX1 and/or IFNGR2 in low-grade gliomas are associated with worse prognosis [9]. In the context of NPC, the EBV-induced latent membrane protein 1 (LMP1) has been shown to elevate PD-L1 expression through the IFNGR2, STAT3, AP-1, and NF-κB pathways [6, 26]. In our study, we identified an increased expression of IFNGR2 in NPC cancer cells. This upregulation of IFNGR2 is implicated in the tumorigenesis of NPC and is positively associated with the expression of PD-L1.
The miRNAs are recognized as potent regulators in the post-transcriptional modulation of gene expression. Through bioinformatics analysis, we pinpointed miR-498-3p as a molecule with potential binding affinity for IFNGR2, and it was found to be consistently underexpressed in NPC cells. Further investigation using luciferase reporter assays substantiated the ability of miR-498-3p to directly interact with IFNGR2. Our findings reveal that an increase in miR-498-3p levels can negatively influence IFNGR2 expression. Previously, miR-498 has been identified to have reduced expression in liver cancer patients and corresponding cell lines, where its overexpression was observed to hinder liver cancer cell proliferation, migration, and invasion, inducing cell cycle arrest and apoptosis [27]. Additional studies have highlighted the role of miR-498 in diminishing colorectal cancer cell proliferation and invasion by targeting Bcl-2 [28], and in mitigating gastric cancer proliferation and chemoresistance via an inverse relationship with Bmi1 [29]. Collectively, these insights suggest miR-498, also named miR-498-3p, functions as a tumor suppressor in the development and progression of NPC cancer. Our data elucidate a novel mechanism wherein miR-498-3p targets IFNGR2 directly, suggesting that overexpression of miR-498-3p could mitigate IFNGR2-mediated repression in NPC.
The circRNAs, as crucial elements of the non-coding RNA family, primarily function by sponging miRNAs to modulate the expression of their target genes [14]. Recent studies have linked the abnormal expression of circRNAs with the progression of various cancers [30, 31]. In the context of NPC, considerable research has been devoted to understanding how circRNAs influence the disease’s pathogenesis. CircRNF13 has been identified as a suppressor of tumor growth and metastasis through its interaction with SUMO2 [32]. Similarly, circRILPL1 is known to activate the Hippo-YAP signaling pathway, thereby facilitating NPC progression [33]. Other circRNAs like circPVT1 and circCRIM1 have been found to enhance NPC metastasis through mechanisms involving the c-Myc/SRSF1 positive feedback loop and the miR-34c-5p/FOSL1 axis, respectively [34, 35]. Given the promising anti-tumor outcomes of PD-1/PD-L1 immune checkpoint therapy, recent investigations have also uncovered the role of various non-coding RNAs, including circRNAs, in modulating PD-1/PD-L1 interactions [36]. For example, circ_002178 has been shown to upregulate PD-L1 expression by sequestering miR-34 in lung adenocarcinoma [37]. Additionally, CircBART2.2 has been reported to regulate PD-L1, contributing to immune evasion in NPC [38]. In a prior investigation, we demonstrated that the circ_0000052/miR-382-3p axis plays a crucial role in upregulating PD-L1 expression in HNSCC [17]. Building on this foundation, our current research leveraged bioinformatics predictions and transcriptome analysis to identify a potential interaction site for miR-498-3p on circ_001377. Subsequent dual luciferase reporter assays confirmed a direct inhibitory relationship between circ_001377 and miR-498-3p. Hsa_circ_001377, also known as hsa_circ_0000054, is mapped to chromosome 1, positions 36920313 to 36923582. This circular RNA is derived from the MRPS15 gene (mitochondrial ribosomal protein S15), a gene that codes for a protein pivotal in mitochondrial translation and protein metabolism. MRPS15 is implicated in vital cellular functions, as evidenced by its gene ontology annotations highlighting its roles in RNA binding and as a structural component of ribosomes. Recent bioinformatics investigations into the mitochondrial ribosomal protein (MRP) family, comprising 82 members, highlighted MRPS15 among five genes showing significant clinical benefits in hepatocellular carcinoma [39]. Additionally, machine learning approaches have pinpointed MRPS15 as one of eight critical mRNA biomarkers in the context of immunoglobulin A nephropathy [40]. Despite these advances, research into the MRP family, particularly the functions of circ_001377 in NPC, remains nascent. We discovered that circ_001377 is elevated in NPC cancer cells and exhibits a positive correlation with PD-L1 expression levels. Interestingly, increasing miR-498-3p levels led to a decrease in IFNGR2 expression. Subsequent experimental manipulations to boost miR-498-3p levels or to augment circ_001377 revealed that elevated circ_001377 levels could competitively interact with miR-498-3p. This interaction diminishes the expression of miR-498-3p, mitigating its suppressive effect on IFNGR2. Consequently, the enhanced IFNGR2 activity triggers the IFN-γ signaling pathway, culminating in the upregulation of PD-L1 in NPC. Our findings unveil a previously unidentified mechanism, by which IFNGR2 may facilitate NPC tumor progression via the interplay between circ_001377 and miR-498-3p, ultimately influencing PD-L1 expression. To our knowledge, this study is the inaugural exploration of IFNGR2’s functionality, establishing the circ_001377/miR-498-3p/ IFNGR2/PD-L1 axis’s regulatory significance in NPC.
CircRNAs are characterized by tissue or cell-specific expressions and unique, highly stable properties, distinguishing them from microRNAs. As such, understanding the role of circRNAs in cancer immunotherapy resistance suggests that targeting these molecules could offer a promising therapeutic strategy for cancer treatment. Approaches like RNAi, antisense technologies, and gene knockout have been used to inhibit oncogenic circRNAs [41]. More recently, the CRISPR-Cas13 system has shown potential for efficiently knocking down circRNAs specifically in cancer cells [42]. Furthermore, studies have demonstrated how circRNAs influence signaling pathways, such as those in glioblastoma [43]. In addition, circRNAs are being explored as potential targets in next-generation RNA vaccines, lipid nanoparticles, exosomes, and virus-like particles. While these delivery methods show promise, they still encounter considerable challenges [41]. Therefore, developing circ_001377-based therapeutics and targeting PD-L1 expression in tumor cells of NPC could offer a promising direction for future treatment strategies.
Conclusions
The present study uncovers the role of IFNGR2 as an oncogenic factor in NPC, where its elevated expression levels are linked to enhance cell proliferation, migration, invasion, clonogenicity, and tumor growth. The interplay between circ_001377 and miR-498-3p contributes to the upregulation of IFNGR2, which enables the activity of IFN-γ signaling pathway, leading to PD-L1 overexpression in NPC. Our findings highlight the circ_001377/miR-498-3p/IFNGR2 axis as a key player in immune evasion mechanisms and propose it as a promising therapeutic target for NPC treatment.
Acknowledgments
None to declare.
Financial Disclosure
This study received funding from the Department of Science and Technology, Jilin Province, China, under the International Science and Technology Cooperation program (grant number: 20220402070GH).
Conflict of Interest
The authors have no conflict of interest to disclose.
Informed Consent
Not applicable.
Author Contributions
GF Guan contributed to the conception of the manuscript, designed and supervised the project, edited and finalized the manuscript. ZM Fu and DJ Zhang: writing the original draft, investigation, methodology, data analysis and interpretation, and conceptualization. YY Guo, F Guo, YN Wan, J Bai, Y Zhao: project administration, methodology, investigation, data collection and analysis. All authors have read and approved the final manuscript.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
| References | ▴Top |
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12(6):431-441.
doi pubmed - Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5(5):423-428.
doi pubmed - Lo KW, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12(6):451-462.
doi pubmed - Chan AT. Current treatment of nasopharyngeal carcinoma. Eur J Cancer. 2011;47(Suppl 3):S302-303.
doi pubmed - Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9-22.
doi pubmed - Johnson D, Ma BBY. Targeting the PD-1/ PD-L1 interaction in nasopharyngeal carcinoma. Oral Oncol. 2021;113:105127.
doi pubmed - Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31(4):539-550.
doi pubmed - Zamora-Salas SX, Macias-Silva M, Tecalco-Cruz AC. Upregulation of the canonical signaling pathway of interferon-gamma is associated with glioblastoma progression. Mol Biol Rep. 2024;51(1):64.
doi pubmed - Zhang X, Chu H, Cheng Y, Ren J, Wang W, Liu X, Yan X. Identification of RUNX1 and IFNGR2 as prognostic-related biomarkers correlated with immune infiltration and subtype differentiation of low-grade glioma. Biomol Biomed. 2023;23(3):405-425.
doi pubmed - Lau TL, Tat-San, Chan LKY, Cheung TH, Yim SF, Lee JHS, Kwong J, et al. Interferon-gamma induces pd-l1 expression via ifngr-jak-stat pathway in ovarian cancer. Cancer Res. 2017;77(13_Supplement):648.
- Ngo J, Chen J, Wu M, Almasan A, Morita H, Jang K-Y, Danielpour D, et al. Interferon gamma receptor 2 (ifngr2) has a ligand (ifng)-independent activity as a bax inhibitor in cancer cells. Cancer Res. 2013;72(8_Supplement): 2013.
- Fijardo M, Yip KW, Liu FF. Mirna biomarkers for npc diagnosis and prognosis. Annls of NPC. 2021;5:1-16.
- Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, Zhang MY, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13(6):633-641.
doi pubmed - Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55-66.
doi pubmed - Yang L, Zhang C, Bai X, Xiao C, Dang E, Wang G. hsa_circ_0003738 inhibits the suppressive function of Tregs by targeting miR-562/IL-17A and miR-490-5p/IFN-gamma signaling pathway. Mol Ther Nucleic Acids. 2020;21:1111-1119.
doi pubmed - Wang WL, Ouyang C, Graham NM, Zhang Y, Cassady K, Reyes EY, Xiong M, et al. microRNA-142 guards against autoimmunity by controlling Treg cell homeostasis and function. PLoS Biol. 2022;20(2):e3001552.
doi pubmed - Zhang DJ, Fu ZM, Guo YY, Guo F, Wan YN, Guan GF. Circ_0000052/miR-382-3p axis induces PD-L1 expression and regulates cell proliferation and immune evasion in head and neck squamous cell carcinoma. J Cell Mol Med. 2023;27(1):113-126.
doi pubmed - Giatromanolaki A, Koukourakis MI, Theodossiou D, Barbatis K, O'Byrne K, Harris AL, Gatter KC. Comparative evaluation of angiogenesis assessment with anti-factor-VIII and anti-CD31 immunostaining in non-small cell lung cancer. Clin Cancer Res. 1997;3(12 Pt 1):2485-2492.
pubmed - Guan G, Zhang D, Zheng Y, Wen L, Yu D, Lu Y, Zhao Y. microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. Int J Clin Exp Pathol. 2014;7(9):5683-5691.
pubmed - Jung HA, Park KU, Cho S, Lim J, Lee KW, Hong MH, Yun T, et al. A phase II study of nivolumab plus gemcitabine in patients with recurrent or metastatic nasopharyngeal carcinoma (KCSG HN17-11). Clin Cancer Res. 2022;28(19):4240-4247.
doi pubmed - Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017;7(2):188-201.
doi pubmed - Refaeli Y, Van Parijs L, Alexander SI, Abbas AK. Interferon gamma is required for activation-induced death of T lymphocytes. J Exp Med. 2002;196(7):999-1005.
doi pubmed - Williams JB, Li S, Higgs EF, Cabanov A, Wang X, Huang H, Gajewski TF. Tumor heterogeneity and clonal cooperation influence the immune selection of IFN-gamma-signaling mutant cancer cells. Nat Commun. 2020;11(1):602.
doi pubmed - Yao RQ, Li ZX, Wang LX, Li YX, Zheng LY, Dong N, Wu Y, et al. Single-cell transcriptome profiling of the immune space-time landscape reveals dendritic cell regulatory program in polymicrobial sepsis. Theranostics. 2022;12(10):4606-4628.
doi pubmed - Slattery ML, Lundgreen A, Bondurant KL, Wolff RK. Interferon-signaling pathway: associations with colon and rectal cancer risk and subsequent survival. Carcinogenesis. 2011;32(11):1660-1667.
doi pubmed - Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, Tang Y, et al. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget. 2014;5(23):12189-12202.
doi pubmed - Zhang X, Xu X, Ge G, Zang X, Shao M, Zou S, Zhang Y, et al. miR-498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep. 2019;41(3):1638-1648.
doi pubmed - Wang T, Ma L, Li W, Ding L, Gao H. MicroRNA-498 reduces the proliferation and invasion of colorectal cancer cells via targeting Bcl-2. FEBS Open Bio. 2020;10(1):168-175.
doi pubmed - Zhao T, Chen Y, Sheng S, Wu Y, Zhang T. Upregulating microRNA-498 inhibits gastric cancer proliferation invasion and chemoresistance through inverse interaction of Bmi1. Cancer Gene Ther. 2019;26(11-12):366-373.
doi pubmed - Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188-206.
doi pubmed - Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42(38):2783-2800.
doi pubmed - Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, Deng X, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20(1):112.
doi pubmed - Wu P, Hou X, Peng M, Deng X, Yan Q, Fan C, Mo Y, et al. Circular RNA circRILPL1 promotes nasopharyngeal carcinoma malignant progression by activating the Hippo-YAP signaling pathway. Cell Death Differ. 2023;30(7):1679-1694.
doi pubmed - Mo Y, Wang Y, Wang Y, Deng X, Yan Q, Fan C, Zhang S, et al. Circular RNA circPVT1 promotes nasopharyngeal carcinoma metastasis via the beta-TrCP/c-Myc/SRSF1 positive feedback loop. Mol Cancer. 2022;21(1):192.
doi pubmed - He W, Zhou X, Mao Y, Wu Y, Tang X, Yan S, Tang S. CircCRIM1 promotes nasopharyngeal carcinoma progression via the miR-34c-5p/FOSL1 axis. Eur J Med Res. 2022;27(1):59.
doi pubmed - Jiang W, Pan S, Chen X, Wang ZW, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer. 2021;20(1):116.
doi pubmed - Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, Kong X, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32.
doi pubmed - Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, Mo Y, et al. Epstein-barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81(19):5074-5088.
doi pubmed - Zhao JW, Zhao WY, Cui XH, Xing L, Shi JC, Yu L. The role of the mitochondrial ribosomal protein family in detecting hepatocellular carcinoma and predicting prognosis, immune features, and drug sensitivity. Clin Transl Oncol. 2024;26(2):496-514.
doi pubmed - Zhang G, Xue L, Zhang S, Liu N, Yao X, Fu J, Nie L. Identification of key biomarkers and signaling pathways and analysis of their association with immune cells in immunoglobulin A nephropathy. Cent Eur J Immunol. 2022;47(3):189-205.
doi pubmed - Ma Y, Wang T, Zhang X, Wang P, Long F. The role of circular RNAs in regulating resistance to cancer immunotherapy: mechanisms and implications. Cell Death Dis. 2024;15(5):312.
doi pubmed - Li S, Li X, Xue W, Zhang L, Yang LZ, Cao SM, Lei YN, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. 2021;18(1):51-59.
doi pubmed - Zhong J, Yang X, Chen J, He K, Gao X, Wu X, Zhang M, et al. Circular EZH2-encoded EZH2-92aa mediates immune evasion in glioblastoma via inhibition of surface NKG2D ligands. Nat Commun. 2022;13(1):4795.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.