Figures
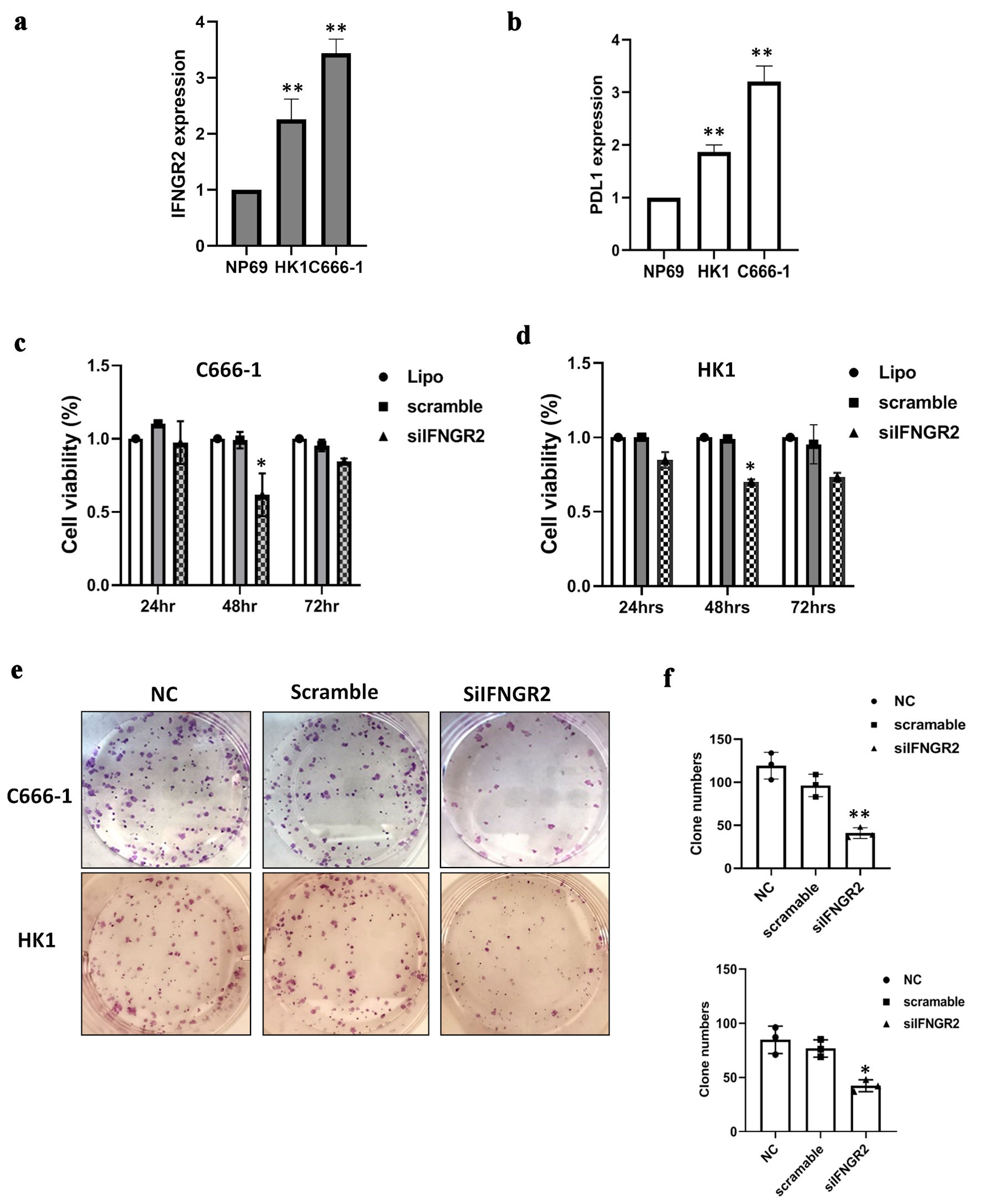
Figure 1. IFNGR2 was upregulated in NPC and promoted cell proliferation and clonogenicity. (a) IFNGR2 overexpression in two NPC cell lines. (b) An increase in PD-L1 levels was noted in NPCs. (c, d) SiRNA targeting IFNGR2 was introduced into HK1 and C666-1 cells, with cell viability assessed at 24, 48, and 72 h post-transfection using the CyQUANT® NF Cell Proliferation Assay. (e, f) Depletion of IFNGR2 significantly reduced the capacity for colony formation. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; NPC: nasopharyngeal carcinoma; PD-L1: programmed cell death-ligand 1; NC: negative control; siRNA: small interfering RNA.
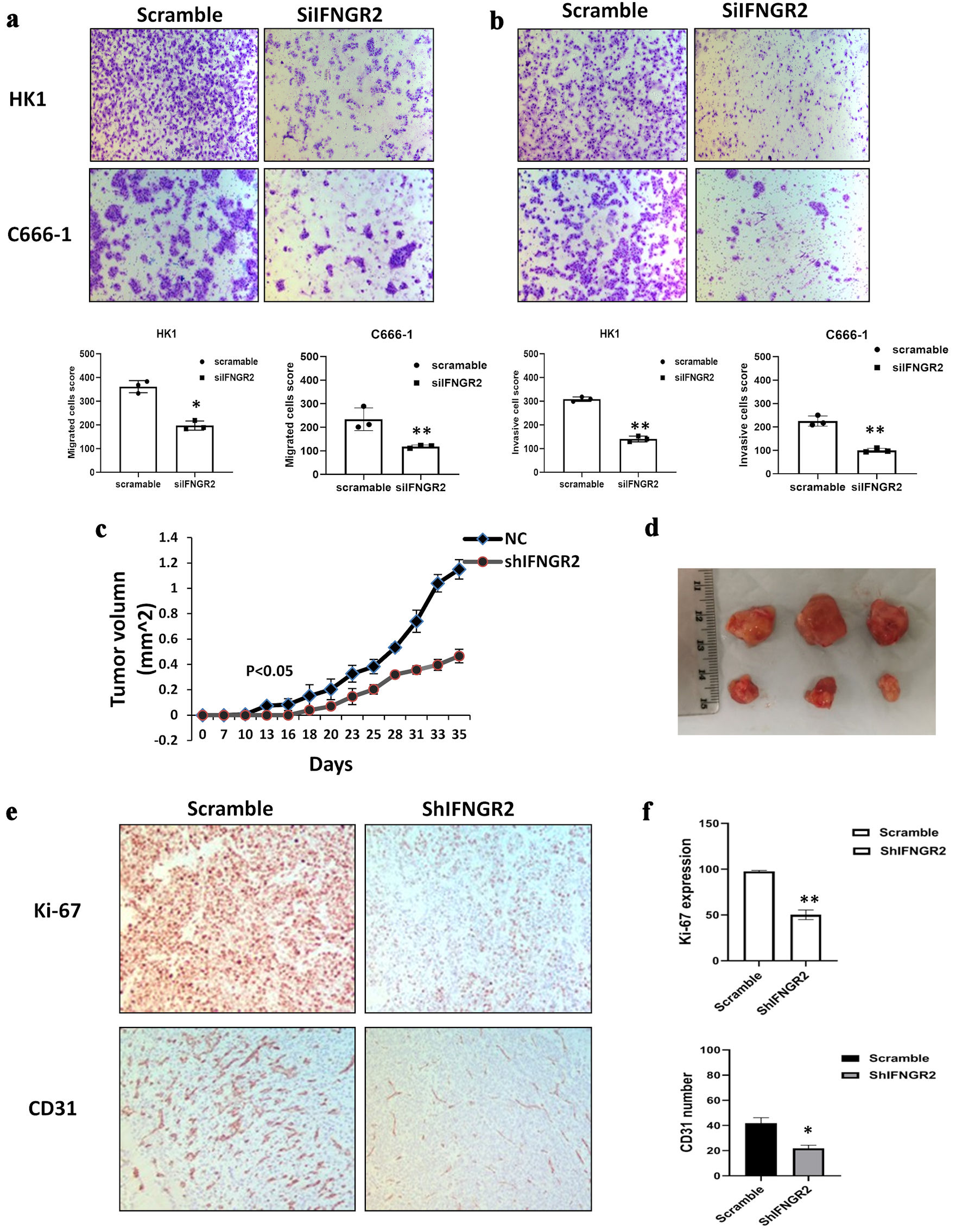
Figure 2. IFNGR2 knockdown inhibited cell migration, invasion and reduced tumor growth in vivo. (a, b) Representative images and quantification of the reduction in migratory capacity (left panel) and invasion (right panel) of HK1 and C666-1 cells that were transfected with IFNGR2-targeting siRNA, compared to negative SC. (c, d) Depletion of IFNGR2 delayed tumor growth compared with the control. Tumor volume was measured. (e, f) Immunohistochemistry detected Ki-67 and CD31 expression in IFNGR2 knockdown group compared with the controls. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; NC: negative control; siRNA: small interfering RNA; SC: scramble control.
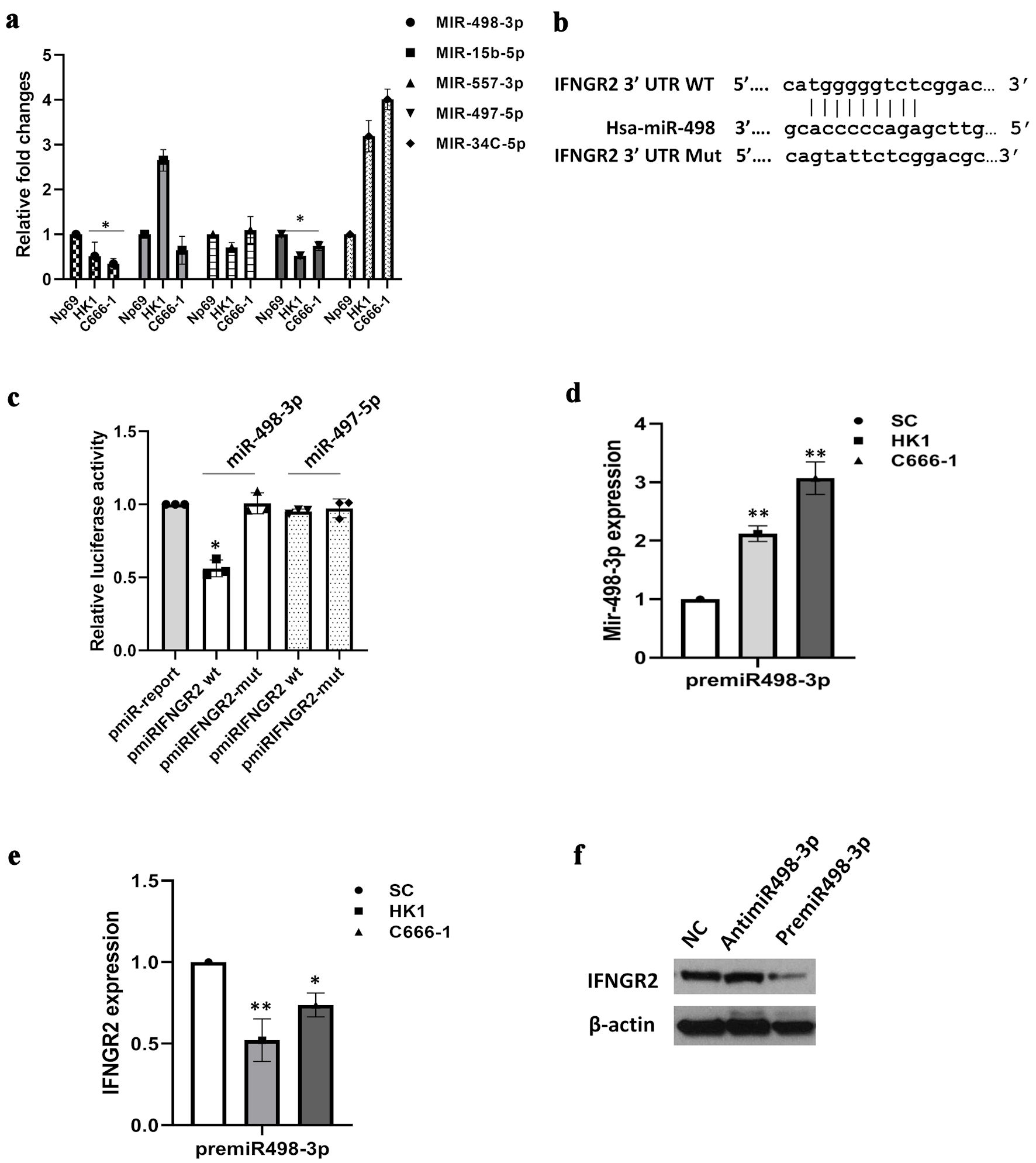
Figure 3. IFNGR2 was directly targeted by miR-498-3p. (a) Bioinformatics analyses led to the identification of five potential miRNA regulators of IFNGR2, with qRT-PCR validations conducted subsequently. (b) Diagram illustrating the binding regions of miR-498-3p on the IFNGR2 sequence, highlighting both wild-type (WT) and mutated (mut) configurations. (c) A dual-luciferase assay verified the specific interaction between miR-498-3p and IFNGR2. (d) Introduction of miR-498-3p mimics elevated miR-498-3p levels in HK1 and C666-1 cells 48 h after transfection. (e) Transfecting cells with premiR-498-3p led to a decrease in IFNGR2 levels. (f) The influence of miR-498-3p on IFNGR2 expression was further confirmed through Western blot analysis. *P < 0.05; **P < 0.01. SC: scramble control; IFNGR2: interferon gamma receptor 2; UTR: untranslated region; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction.
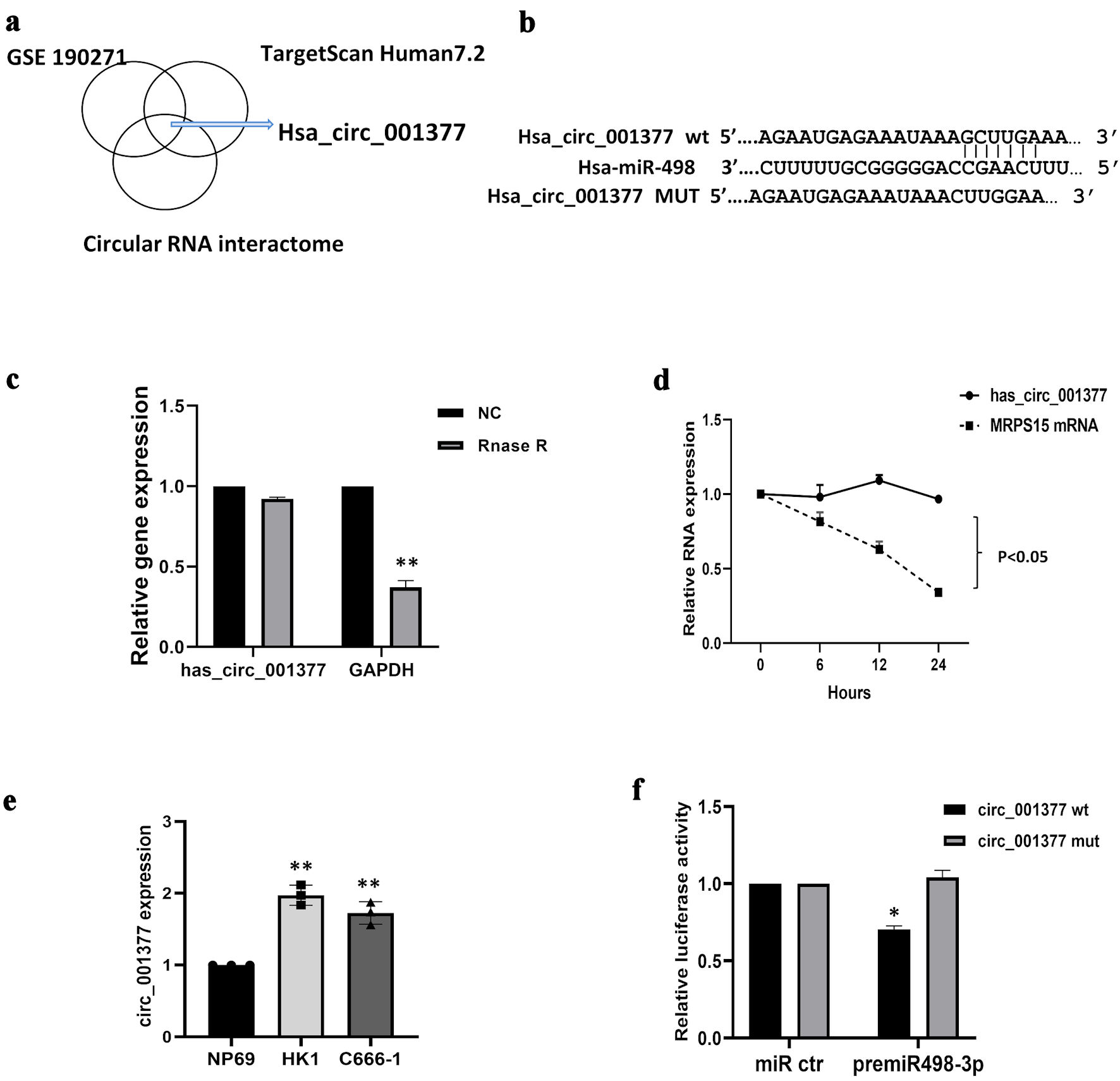
Figure 4. Circ_001377 acted as a sponge for miR-498-3p. (a) Circ_001377 identification involved circular RNA interaction predictors, TargetScan Human7.2, and GSE190271 dataset analysis, complemented by KEGG pathway analysis. (b) Diagram depicting the miR-498-3p binding site on the wild-type (WT) circ_001377 sequence alongside a mutant variant of circ_001377. (c) Following RNase R or mock treatment of C666-1 cell-derived RNA, circ_001377 stability was assessed via qRT-PCR. (d) C666-1 cells were treated with actinomycin D, the relative mRNA levels were measured by qRT-PCR. (e) Circ_001377 expression was measured across NPC cell lines. (f) Luciferase assay outcomes in C666-1 cells transfected with luciferase reporters linked to miR-498-3p targets on WT or mutated circ_001377 sites, alongside miR-498-3p mimics or controls. *P < 0.05; **P < 0.01. NC: negative control; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction; NPC: nasopharyngeal carcinoma.
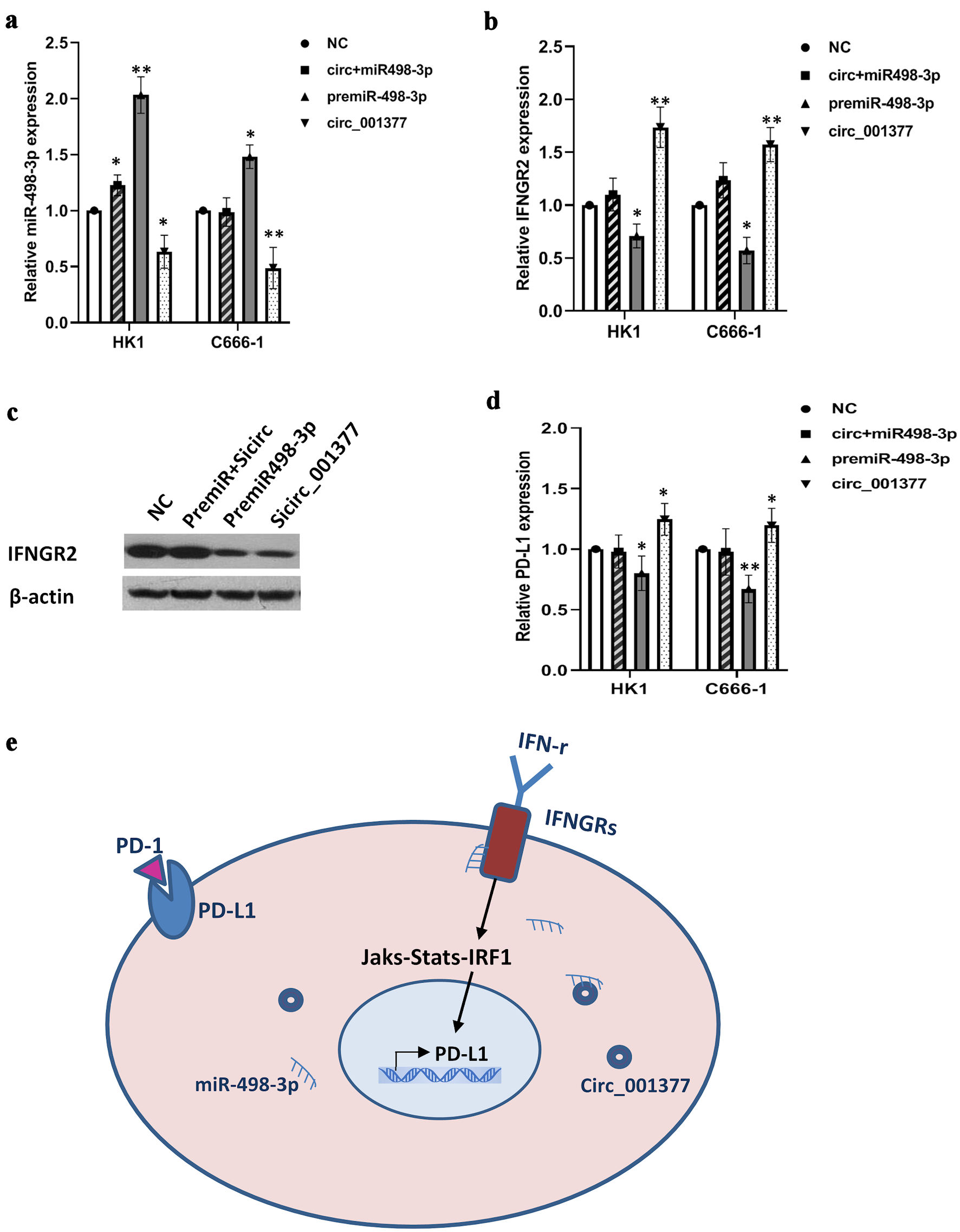
Figure 5. Circ_001377 bound to miR-498-3p, reducing its interaction with IFNGR2 and regulating PD-L1 expression. (a, b) Rescue experiments conducted with premiR-498-3p, circ_001377 overexpression plasmids, or a combination thereof, followed by qRT-PCR assessment of miR-498-3p and IFNGR2. (c) Alterations in IFNGR2 expression caused by miR-498-3p enhancement or circ_001377 depletion were assessed by Western blotting. (d) PD-L1 expression was influenced by the introduction of premiR-498-3p, overexpression plasmids for circ_001377, or a combination of both. (e) Schematic of a proposed model where circ_001377 functions as a sponge to sequester miR-498-3p, resulting in increased IFNGR2 levels. This, in turn, enhanced the IFN-γ signaling pathway, leading to the overexpression of PD-L1 in NPC. *P < 0.05; **P < 0.01. IFNGR2: interferon gamma receptor 2; qRT-PCR: quantitative reverse transcriptase-polymerase chain reaction; NC: negative control; PD-L1: programmed cell death-ligand 1; IFN-γ: interferon gamma; PD-1: programmed cell death protein-1.




