| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://wjon.elmerpub.com |
Original Article
Volume 16, Number 1, February 2025, pages 104-112
Methylation of SOX1 and PAX1 Are Risk Factors and Potential Biomarkers for Cervical Lesions
Yan Die Lina, b, Xiao Yue Lia, b, Li Wei Shaoa, Ai Jun Liua, c
aDepartment of Pathology, The Seventh Medical Center, Chinese PLA General Hospital, Beijing 100700, China
bThese authors contributed equally to this work.
cCorresponding Author: Ai Jun Liu, Department of Pathology, The Seventh Medical Center, Chinese PLA General Hospital, Beijing 100700, China
Manuscript submitted October 3, 2024, accepted December 10, 2024, published online December 31, 2024
Short title: Methylation of SOX1 and PAX1 and Cervical Lesions
doi: https://doi.org/10.14740/wjon1985
| Abstract | ▴Top |
Background: The correlation between methylation of paired box gene 1 (PAX1) and sex determining region Y-box 1 (SOX1) with human papillomavirus (HPV) infection and the progression of cervical lesions is not well understood. This study aims to explore the potential value of PAX1 and SOX1 as diagnostic biomarkers for cervical diseases.
Methods: A total of 139 cervical biopsy tissue samples were obtained from the Department of Pathology, the Seventh Medical Center, Chinese PLA General Hospital from 2021 to 2023. The samples include 32 cases of chronic cervicitis (inflammation group), 30 cases of low-grade squamous intraepithelial lesions (LSIL group), 50 cases of high-grade squamous intraepithelial lesions (HSIL group), and 27 cases of cervical squamous cell carcinoma (CSCC group). DNA was extracted from paraffin-embedded tissues, and the levels of HPV infection and methylation of PAX1 and SOX1 were detected.
Results: The methylation index (M-index) of PAX1 and SOX1 in the HSIL and CSCC groups is significantly higher than in the inflammation group (both P < 0.0001), with no significant difference between the LSIL and inflammation groups. There is no significant difference in the positive PAX1 and SOX1 methylation rate with HPV infection and age. The positive rates of PAX1 methylation in the inflammation, LSIL, HSIL, and CSCC groups were 3.13%, 10.00%, 44.00%, and 88.89%, respectively. The positive rates of SOX1 methylation were 3.13%, 10.00%, 40.00%, and 77.78%, respectively, and increasing with the progression of cervical lesions (R2 = 0.9189/R2 = 0.9279, P < 0.0001/P < 0.0001). Comparing LSIL, HSIL, and CSCC with the inflammation group and using cervical biopsy pathology diagnosis as the gold standard, methylation of PAX1 and SOX1 is a risk factor for HSIL and CSCC, with odds ratio (OR) values significantly increasing as lesions progress. The sensitivity of PAX1 and SOX1 methylation to cervical lesions increases with the progression of the lesions.
Conclusions: Methylation of SOX1 and PAX1 is not associated with HPV infection. The positive rate of methylation for SOX1 and PAX1 is positively correlated with cervical lesions, which can serve as potential biomarkers for HSIL and CSCC. They are risk factors and potential screening indicators for HSIL and above cervical lesions.
Keywords: Cervical squamous cell carcinoma; Methylation; PAX1; SOX1; HPV
| Introduction | ▴Top |
Cervical cancer (CC) is one of the most common malignancies among women worldwide, with a high incidence and mortality rate. It ranks fourth among causes of cancer-related deaths in women, thereby remaining a significant threat to female health [1]. Persistent infection with high-risk human papillomavirus (HPV) has been identified as the primary cause of CC [2]. The traditional screening methods are Pap smears, HPV testing, and colposcopy. Recent studies have suggested the role of DNA methylation alterations in the progression of CC, indicating that DNA methylation can serve as an effective biomarker for CC screening [3]. Studies have also shown that DNA methylation can be used as an effective biomarker for the diagnosis of CC [4].
Methylation of DNA involves the addition of a methyl group to the cytosine nucleotide in DNA, under the influence of DNA methyltransferases (DNMTs), leading to the formation of 5-methylcytosine. More than half of the genes in the genomes of vertebrate contain short regions rich in CG dinucleotides, known as CpG islands (CGIs), which are the primary sites of methylation [5]. The PAX gene family, named after the paired box DNA-binding domain, plays a crucial role in tissue development and cell differentiation during embryogenesis [6]. The findings demonstrated that the expression of SOX1 is associated with early embryogenesis, development of the central nervous system, and maintenance of neural stem cells [7]. SOX1 and PAX1 are tumor suppressor genes that regulate processes related to cell proliferation and invasion [8, 9], Their methylation occurs at a high frequency in CC and significantly increases as the disease progresses [10]. However, current studies are mostly focused on the detection of cervical exfoliated cells, with fewer investigations on the use of PAX1 and SOX1 methylation in cervical tissue for the diagnosis of CC. The purpose of this study is to explore the potential value of PAX1 and SOX1 methylation as diagnostic biomarkers for CC.
| Materials and Methods | ▴Top |
Study population and data collection
A total of 139 cervical biopsy tissues were collected from the Seventh Medical Center of PLA General Hospital, including 32 cases of chronic cervicitis, 30 cases of low-grade squamous intraepithelial lesion (LSIL), 50 cases of high-grade squamous intraepithelial lesion (HSIL), and 27 cases of cervical squamous cell carcinoma (CSCC). The inclusion criteria are as follows: 1) female patients aged between 20 and 75 years; 2) patients who underwent cervical biopsy under colposcopic examination, with the biopsy slides reviewed and diagnosed by two pathologists; 3) exclusion of specimens from patients with other malignant tumors, immunodeficiency diseases, other cervical lesions, or those who were currently pregnant. The clinical characteristics of the patients are provided in Table 1.
 Click to view | Table 1. The Clinical Characteristics of the 139 Patients |
HPV DNA testing
HPV DNA testing in paraffin-embedded tissue samples blocks containing ≥ 20% lesions were selected, and DNA from tissue samples was extracted using the formalin-fixed paraffin-embedded (FFPE) tissue extraction kit (AmoyDx, China). The concentration and purity of the DNA samples were evaluated using Nanodrop (Thermo Fisher Scientific). HPV genotyping was performed using an HPV test kit (Kapa Biosystems, China) that can identify 21 HPV genotypes, including 15 high-risk HPV DNAs (types 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, and 68) and six low-risk HPV DNAs (types 6, 11, 42, 43, 44, and 81).
Methylation testing for PAX1 and SOX1
After extracting DNA from tissue samples, bisulfite conversion was performed, and then the methylation status was assessed by amplifying the converted DNA using multiplex polymerase chain reaction (PCR). DNA was extracted from paraffin-embedded tissues (Aidlab Biotechnologies, nucleic acid extraction reagent) and converted through a chemical reaction with bisulfite, which transforms unmethylated cytosine into uracil sulfonate through a deamination reaction. Multiplex PCR amplification was utilized to determine the bisulfite-converted DNA, thus detecting the methylation of SOX1, PAX1, and the internal control gene ACTB (Kapa Pharmaceuticals, methylation testing kits for SOX1 and PAX1 genes). The positive cut-off value for PAX1 was 38, and for SOX1, it was 38.6. The formula for the methylation index (M-index) is M-index = 10,000 × 2-|Ct value of target gene - Ct value of internal control gene| [11].
Statistical analyses
The M-index is presented as mean ± standard deviation. A comparison between two groups was performed using the two-independent sample t-test. Data analysis was conducted using SPSS statistical software (version 21.0), and GraphPad Prism 8 was used for data visualization and statistical plotting. P value < 0.05 was considered statistically significant.
Ethics approval
This study was approved by the Ethics Committee of the Seventh Medical Center of Chinese PLA General Hospital (approval number: 2022-181) and was conducted in compliance with the ethical standards of the responsible institution on human subjects as well as with the Helsinki Declaration.
| Results | ▴Top |
A total of 139 patients were enrolled in this study from 2020 to 2023. These patients were divided into four groups: 32 in the inflammation group, with an average age of 45.40 ± 15.32 years; 30 in the LSIL group, with an average age of 41.16 ± 11.68 years; 50 in the HSIL group, with an average age of 40.62 ± 9.86 years; and 27 in the CSCC group, with an average age of 53.37 ± 9.05 years. The results revealed no significant difference between the two groups in terms of age. All patients were diagnosed by clinical pathologists. Representative hematoxylin and eosin (H&E) stained images of cervical biopsy tissues are shown in Figure 1.
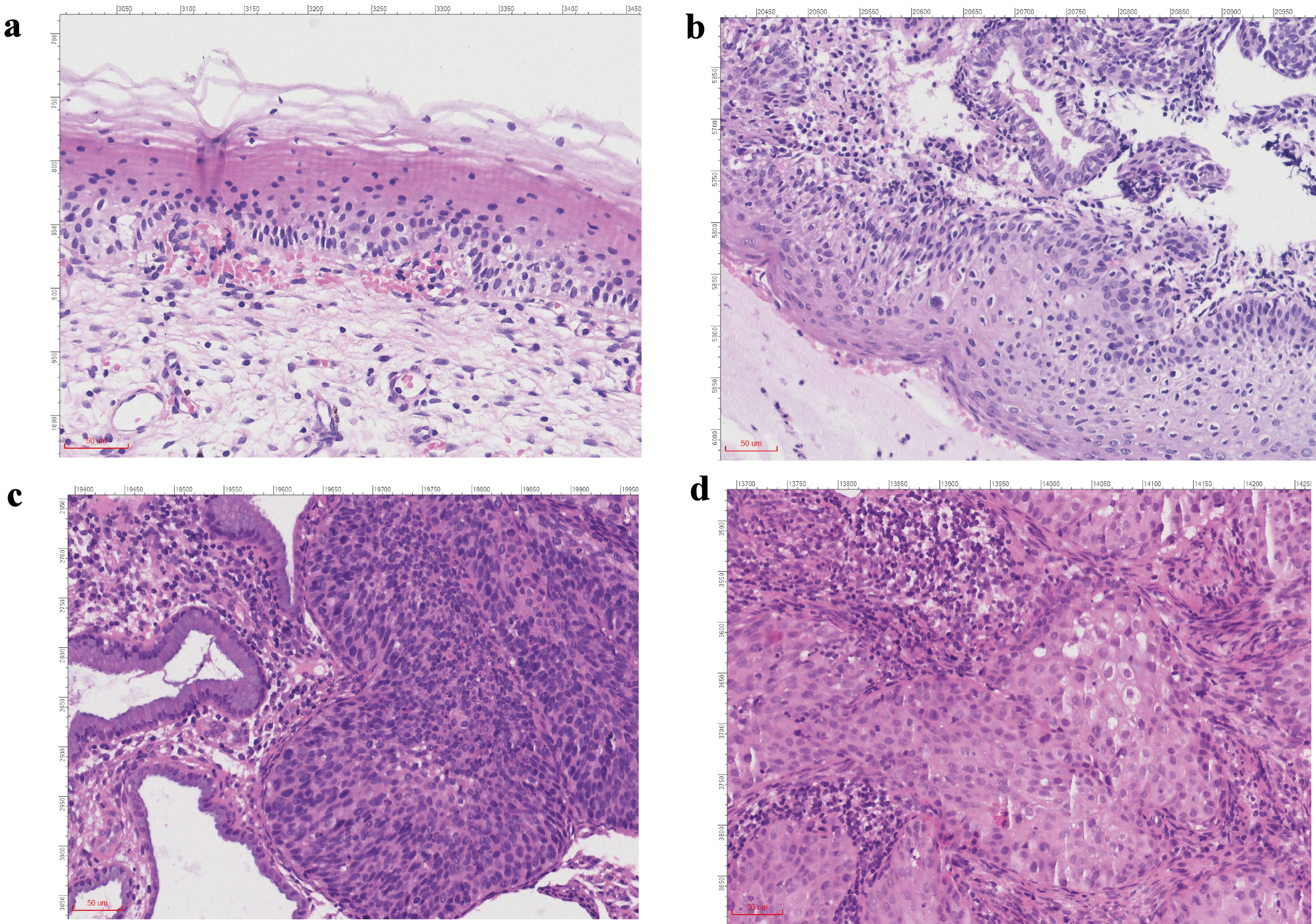 Click for large image | Figure 1. H&E staining images of patients in each group (× 100). (a) Inflammation group. (b) LSIL group. (c) HSIL group. (d) CSCC group. H&E: hematoxylin and eosin stain; LSIL: low-grade squamous intraepithelial lesion; HSIL: high-grade squamous intraepithelial lesion; CSCC: cervical squamous cell carcinoma. |
Analysis of PAX1 and SOX1 methylation indices in cervical lesion tissues
We analyzed the methylation indices of PAX1 and SOX1 in 139 cervical biopsy tissue samples. The results indicated minimal methylation in the inflammation group, with median and quartile values of 0 for both PAX1 and SOX1. In the LSIL group, the methylation values of PAX were 0. Similarly, for SOX1, the median and quartile values were also 0. There was no significant difference between the LSIL group and the inflammation group (P > 0.05) In the HSIL group, the methylation indices for PAX1 and SOX1 were 0.9150 and 36.21, showing a significant increase compared to the inflammation group (P < 0.0001). In the CSCC group, the methylation indices dramatically increased to 1,259 for PAX1 and 1,743 for SOX1, which showed the highest values across all groups (Fig. 2, Table 2). These results indicate a significant increase in the M-Index of PAX1 and SOX1 in the HSIL and CSCC groups, highlighting a trend where the methylation levels increase progressively as cervical lesions advance.
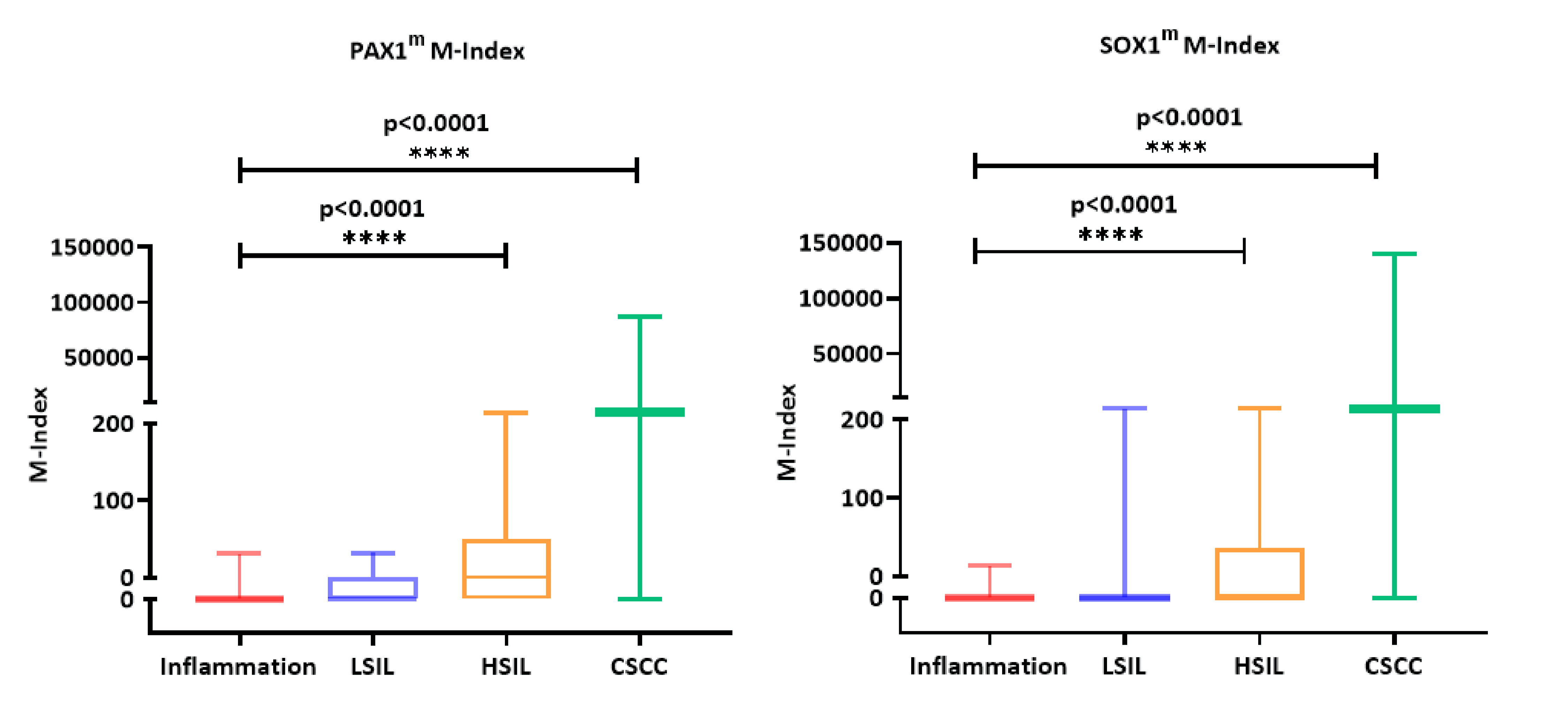 Click for large image | Figure 2. Methylation index of PAX1 (a) and SOX1 (b) in paraffin-embedded tissue samples of 139 patients undergoing cervical biopsy. LSIL: low-grade squamous intraepithelial lesion; HSIL: high-grade squamous intraepithelial lesion; CSCC: cervical squamous cell carcinoma; M-index: methylation index; PAX1m: PAX1 methylation; SOX1m: SOX1 methylation. |
 Click to view | Table 2. Methylation Index of PAX1 and SOX1 |
The increased positive rate of SOX1 and PAX1 methylation during the progression of CC
We converted the quantitative Ct values into qualitative data and analyzed the methylation positive rates using 38 and 36 as the positive cut-off values for PAX1 and SOX1, respectively. In the inflammation group, the positive rates for both SOX1 and PAX1 were 3.13%, with a concurrent positive rate for both SOX1 and PAX1 being 6.25%. In the LSIL group, the results showed that the positive rates for both SOX1 and PAX1 were 10%, with a concurrent positive rate of 13.33%. The positive rates for both SOX1 and PAX1 were 44% and 40% in the HSIL group, with a concurrent positive rate reaching 52%. In the CSCC group, the results showed that the positive rates for both SOX1 and PAX1 increased to 88.89% and 77.78%, respectively, with a concurrent positive rate reaching 88.89%. The research findings demonstrate that the positive rates of methylation for PAX1 (R2 = 0.9189), SOX1 (R2 = 0.9279), or both (R2 = 0.9367) are positively associated with the progression of cervical lesions (P < 0.01) (Fig. 3a, Table 3). The trend is consistent in both single-gene (PAX1 or SOX1) and dual-gene (PAX1 and SOX1) analyses, with the positive rates for the dual-gene analysis slightly higher than those for the single-gene tests. Furthermore, the results of correlation analysis indicated that, within the diseased cervical tissues, there was no significant association between PAX1/SOX1 methylation and either different age groups or HPV infection status (Fig. 3b, c, Table 3).
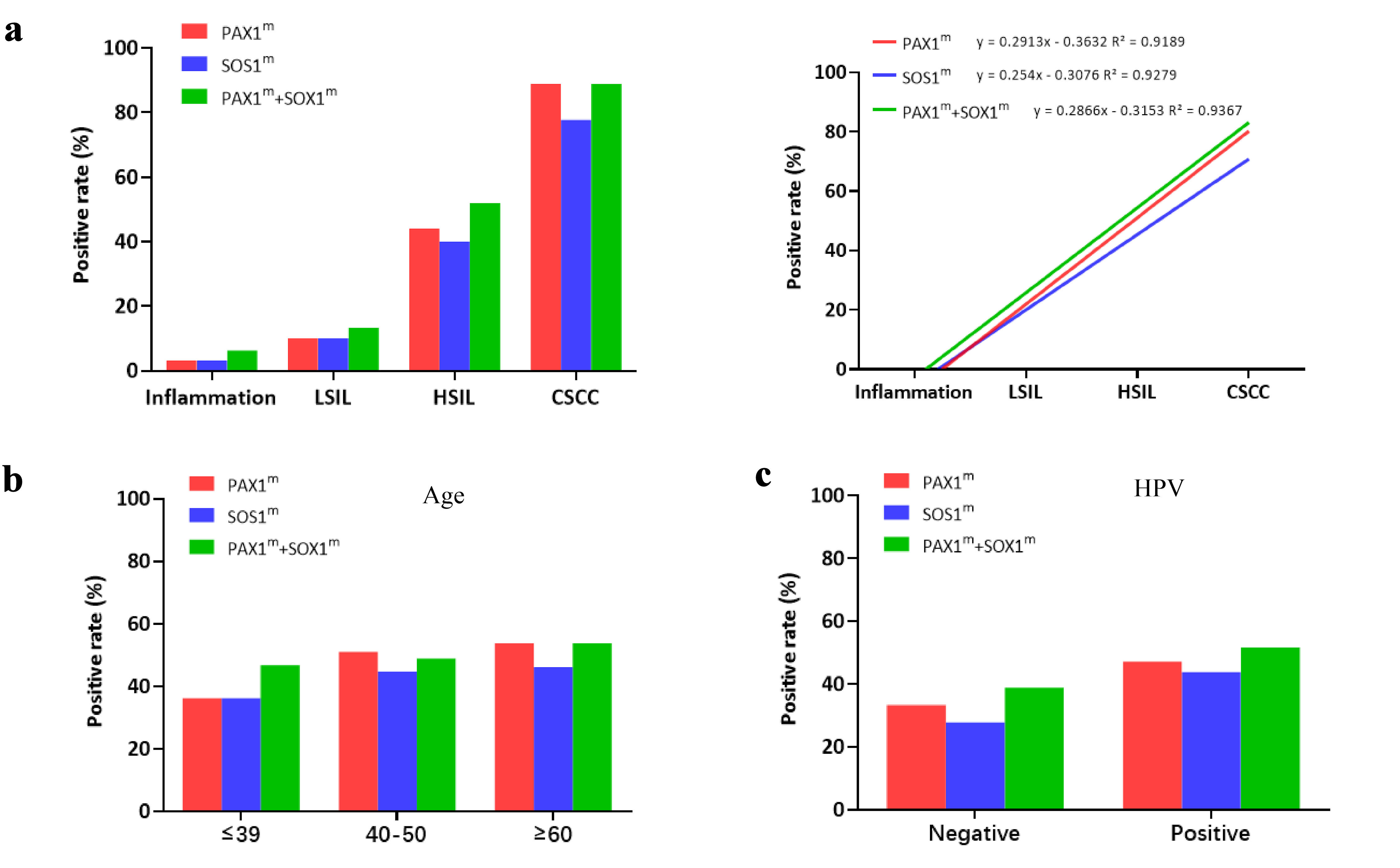 Click for large image | Figure 3. Methylation positivity rates of PAX1 and SOX1 in paraffin-embedded tissue samples of 139 patients undergoing cervical biopsy. (a) Methylation positivity rates of PAX1 and SOX1 in cervical biopsy tissue samples of patients in different stages. (b) Methylation positivity rates of PAX1 and SOX1 in cervical biopsy tissue samples of patients in different age groups. (c) Methylation positivity rates of PAX1 and SOX1 in cervical biopsy tissue samples of patients with HPV infection. LSIL: low-grade squamous intraepithelial lesion; HSIL: high-grade squamous intraepithelial lesion; CSCC: cervical squamous cell carcinoma; HPV: human papillomavirus; M-index: methylation index; PAX1m: PAX1 methylation; SOX1m: SOX1 methylation. |
 Click to view | Table 3. Methylation Positivity Rates of PAX1 and SOX1 in Cervical Biopsy Tissue Samples of 139 Patients |
Evaluating the efficacy of HPV testing, PAX1, and SOX1 methylation in identifying cervical lesions
We first evaluated the diagnostic efficacy of HPV for cervical diseases, and the results showed that HPV was identified as a risk factor for LSIL, HSIL, and CSCC (Table 4). Subsequently, we evaluated the efficacy of PAX1 and SOX1 methylation in identifying cervical lesions. In the LSIL group, the specificity of the methylation tests for both PAX1 and SOX1 genes is 96.88%, but the sensitivity and odds ratio (OR) are relatively low, being 10% and 3.44 respectively. The sensitivity of PAX1 combined with SOX1 is only 13.33%, with an OR of 2.31 (P = 0.35, 95% confidence interval (CI): 0.39 - 13.64), suggesting that PAX1 and SOX1 methylation may not be significant risk factors for disease in the LSIL group. However, in the HSIL and CSCC groups, the sensitivity of the combined PAX1 and SOX1 gene methylation detection was 52.00% and 88.89%, respectively. The specificity of both groups was 93.75%, and ORs of 16.25 (P < 0.0001, 95% CI: 18.53 - 777.02) and 120.00 (P < 0.0001, 95% CI: 3.50 - 75.43) (Table 4). These results indicate that methylation testing of PAX1 and SOX1 genes has the potential to serve as a risk biomarker for patients with HSIL and higher grades of cervical lesions.
 Click to view | Table 4. Sensitivity, Specificity, and Odds Ratios (ORs) of HPV Detection, PAX1 Methylation Detection, and SOX1 Methylation Detection in 139 Patients |
| Discussion | ▴Top |
CC is one of the most prevalent malignancies among females. Persistent infection by high-risk HPV types can lead to precancerous lesions or even CC. The combined testing of HPV and liquid-based cytology has been effective in preventing the occurrence and progression of CC [12]. However, traditional detection methods are subjective and lack precision, so there is an urgent need for more accurate methods. DNA methylation is an epigenetic modification and is considered a novel biomarker for the detection of CC. Previous studies have shown that methylation of the PAX1 gene can be used as an auxiliary to routine cytology in CC screening [13]. Some studies have also confirmed the correlation between DNA methylation levels of genes such as PAX1, SOX1, LMX1A, and ZNF582 and the occurrence and progression of CC [14, 15]. Nevertheless, the potential value of DNA methylation in CC screening or early diagnosis has not yet been fully elucidated. Large-scale studies are needed to further explore the potential methylation biomarkers for cervical lesions. PAX1 is a tumor suppressor gene, and its high level of methylation is associated with the occurrence of tumors, which has been identified in cancers such as ovarian and oral cancers [16, 17]. The research results of Su et al suggest that PAX1 can activate a variety of phosphatases, thereby regulating oncogenic kinases and exerting an effect on tumor growth and proliferation in cellular signal transduction [18]. The methylation of SOX1 has been reported to be associated with various types of cancer, including liver cancer, lung cancer, and urothelial carcinoma [19-21]. In this study, by using the M-index as a measure, our results showed that the M-index for PAX1 and SOX1 was significantly higher in HSIL and CSCC than in the inflammation group, but there was no statistical difference in LSIL group compared to the inflammation group. These findings suggest that PAX1 and SOX1 methylation may play an important role in the progression of cervical lesions to HSIL and CSCC stages.
According to literature reports, in samples of cervical exfoliated cells, high methylation of PAX1 positively correlates with high HPV viral load [22]. However, it has been reported that gene methylation is associated with the progression of CC, but there is no significant correlation with HPV infection [23]. In this study, we assessed HPV in cervical tissue lesions to explore its correlation with methylation of PAX1/SOX1. Our findings suggest that HPV infection is not associated with PAX1/SOX1 methylation. These results suggest that methylation of the SOX1 and PAX1 genes may be an independent factor in the development of CC. In addition, as the severity of cervical lesions increases, the positive rates of PAX1 and SOX1 methylation correspondingly increase, reaching the highest in CSCC. Zhang et al reported methylation positivity rates of PAX1 and SOX1 in LSIL at 43.7% and 19.6%, and in HSIL at 68.8% and 55.9%, respectively [10]. Huang et al noted the SOX1 methylation rate exhibited a significant upward trend (P < 0.001), escalating from 27.27% in LSIL samples to 40.75% in HSIL samples, and culminating at 84.56% in CC samples [24]. Xu et al reported a methylation positivity rate of PAX1 in LSIL samples at 9%, and in HSIL at 44% [25]. Compared to previous reports, our findings reveal discrepancies in PAX1 and SOX1 methylation positivity rates in LSIL but a higher consistency in HSIL and CSCC. These findings suggest that the methylation of SOX1 and PAX1 may have a potential role in the later stages of cervical lesion development.
Currently, cytology and HPV testing have been widely used for CC screening. Patients diagnosed with HSIL require subsequent treatment, including ablation or cone biopsy [26]. However, about 10-24% of women diagnosed with HSIL who undergo loop electrosurgical excision procedure (LEEP) have only minor and mild lesions [27, 28], and this procedure is accompanied by complications including infertility, premature birth, and premature rupture of membranes. Therefore, for LSIL patients who are at risk of progressing to more serious diseases, more precise treatment strategies are needed. The p16/Ki-67 immunohistochemistry has emerged as a potential biomarker with high sensitivity and specificity for identifying HSIL [29]. However, this type of detection has a limitation in subjectivity. Therefore, exploring a reliable molecular biomarker for triage is particularly important. The results of a meta-analysis of seven studies involving 1,055 patients showed that PAX1 methylation is associated with the transition from normal cervical epithelium to cervical intraepithelial neoplasia and cancer, which indicates its potential value as a biomarker for cervical diseases [30]. In the study, we revealed the potential value of PAX1 and SOX1 methylation in the diagnosis of cervical lesions, particularly in the identification of high-grade lesions. In the LSIL group, although methylation detection of PAX1 and SOX1 showed high specificity, the sensitivity and OR values were relatively low, indicating that these methylation markers may not be sufficient as independent diagnostic tools in the early stages of cervical lesions. This finding is consistent with previous studies [31, 32], suggesting that at the LSIL stage, it may be necessary to combine other biomarkers or diagnostic methods to enhance the sensitivity of detection. However, in the HSIL and CSCC groups, methylation detection of PAX1 and SOX1 exhibited higher sensitivity and specificity, along with a significantly increased OR value, emphasizing the diagnostic value of these markers in the later stages of cervical lesions. These results highlight the potential value of PAX1 and SOX1 methylation as diagnostic biomarkers for CC, but its clinical applicability requires further investigation to validate its effectiveness. Our study also has certain limitations. Firstly, our study is limited to patients with cervical squamous cell carcinoma and does not include cervical adenocarcinoma, the effects of PAX1 and SOX1 methylation on cervical adenocarcinoma remain to be further validated. Additionally, our research sample size is limited, and more data and clinical follow-up are still needed for further exploration.
Conclusions
In conclusion, our study indicates that PAX1 and SOX1 methylation significantly increase HSIL and reach the highest levels in CSCC. As cervical lesions progress, the positive rates of SOX1 and PAX1 methylation gradually increase, showing good sensitivity and specificity for late-stage cervical lesions and serving as potential biomarkers for higher-grade cervical lesions. The findings of this study provide a theoretical foundation for the use of methylation markers in the clinical diagnosis of CC. However, further research is required to explore their broader application in clinical practice testing in the clinical diagnosis of CC.
Acknowledgments
None to declare.
Financial Disclosure
This work was supported by the 2023 Innovation Cultivation Fund of the Seventh Medical Center of the Chinese PLA General Hospital (No. qzx-2023-13).
Conflict of Interest
The authors declare no conflict of interest.
Informed Consent
Written informed consents were obtained from the patients for publication.
Author Contributions
YDL: writing - review and editing, writing - original draft, formal analysis, data curation. XYL: formal analysis, data curation. LWS: investigation, conceptualization, methodology. AJL: validation, supervision, conceptualization, writing - review and editing.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
| References | ▴Top |
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249.
doi pubmed - Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889-899.
doi pubmed - Wright TC, Jr., Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D, American Society for C, et al. 2006 consensus guidelines for the management of women with abnormal cervical cancer screening tests. Am J Obstet Gynecol. 2007;197(4):346-355.
doi pubmed - Shivapurkar N, Sherman ME, Stastny V, Echebiri C, Rader JS, Nayar R, Bonfiglio TA, et al. Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecol Oncol. 2007;107(3):549-553.
doi pubmed - Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484-492.
doi pubmed - Robson EJ, He SJ, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6(1):52-62.
doi pubmed - Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269(2):580-594.
doi pubmed - Tsao CM, Yan MD, Shih YL, Yu PN, Kuo CC, Lin WC, Li HJ, et al. SOX1 functions as a tumor suppressor by antagonizing the WNT/beta-catenin signaling pathway in hepatocellular carcinoma. Hepatology. 2012;56(6):2277-2287.
doi pubmed - Maulbecker CC, Gruss P. The oncogenic potential of Pax genes. EMBO J. 1993;12(6):2361-2367.
doi pubmed - Zhang L, Yu J, Huang W, Zhang H, Xu J, Cai H. A sensitive and simplified classifier of cervical lesions based on a methylation-specific PCR assay: a Chinese cohort study. Cancer Manag Res. 2020;12:2567-2576.
doi pubmed - Huang YK, Peng BY, Wu CY, Su CT, Wang HC, Lai HC. DNA methylation of PAX1 as a biomarker for oral squamous cell carcinoma. Clin Oral Investig. 2014;18(3):801-808.
doi pubmed - Sun XF, Gu YQ, Wang AC, Wang J, Xie JL. [Value assessment of high-risk HPV test and TCT in the screening of cervical carcinoma]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2013;27(4):273-276.
pubmed - Hsu YW, Huang RL, Su PH, Chen YC, Wang HC, Liao CC, Lai HC. Genotype-specific methylation of HPV in cervical intraepithelial neoplasia. J Gynecol Oncol. 2017;28(4):e56.
doi pubmed - Rogeri CD, Silveira HCS, Causin RL, Villa LL, Stein MD, de Carvalho AC, Arantes L, et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol Oncol. 2018;150(3):545-551.
doi pubmed - van Leeuwen RW, Ostrbenk A, Poljak M, van der Zee AGJ, Schuuring E, Wisman GBA. DNA methylation markers as a triage test for identification of cervical lesions in a high risk human papillomavirus positive screening cohort. Int J Cancer. 2019;144(4):746-754.
doi pubmed - Su HY, Lai HC, Lin YW, Chou YC, Liu CY, Yu MH. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int J Cancer. 2009;124(2):387-393.
doi pubmed - Guerrero-Preston R, Michailidi C, Marchionni L, Pickering CR, Frederick MJ, Myers JN, Yegnasubramanian S, et al. Key tumor suppressor genes inactivated by "greater promoter" methylation and somatic mutations in head and neck cancer. Epigenetics. 2014;9(7):1031-1046.
doi pubmed - Su PH, Lai HC, Huang RL, Chen LY, Wang YC, Wu TI, Chan MWY, et al. Paired Box-1 (PAX1) activates multiple phosphatases and inhibits Kinase cascades in cervical cancer. Sci Rep. 2019;9(1):9195.
doi pubmed - Liu XY, Fan YC, Gao S, Zhao J, Chen LY, Li F, Wang K. Methylation of SOX1 and VIM promoters in serum as potential biomarkers for hepatocellular carcinoma. Neoplasma. 2017;64(5):745-753.
doi pubmed - Li N, Li X, Li S, Zhou S, Zhou Q. Cisplatin-induced downregulation of SOX1 increases drug resistance by activating autophagy in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2013;439(2):187-190.
doi pubmed - Lopez JI, Angulo JC, Martin A, Sanchez-Chapado M, Gonzalez-Corpas A, Colas B, Ropero S. A DNA hypermethylation profile reveals new potential biomarkers for the evaluation of prognosis in urothelial bladder cancer. APMIS. 2017;125(9):787-796.
doi pubmed - Li M, Zhao C, Zhao Y, Li J, Zhang X, Zhang W, Gao Q, et al. Association and effectiveness of PAX1 methylation and HPV viral load for the detection of cervical high-grade squamous intraepithelial lesion. Pathogens. 2022;12(1):63.
doi pubmed - Jha AK, Nikbakht M, Jain V, Capalash N, Kaur J. p16(INK4a) and p15(INK4b) gene promoter methylation in cervical cancer patients. Oncol Lett. 2012;3(6):1331-1335.
doi pubmed - Huang J, Gao H, Tan HZ. SOX1 promoter hypermethylation as a potential biomarker for high-grade squamous intraepithelial neoplasia lesion and cervical carcinoma: a meta-analysis with trial sequential analysis. Front Genet. 2020;11:633.
doi pubmed - Xu J, Xu L, Yang B, Wang L, Lin X, Tu H. Assessing methylation status of PAX1 in cervical scrapings, as a novel diagnostic and predictive biomarker, was closely related to screen cervical cancer. Int J Clin Exp Pathol. 2015;8(2):1674-1681.
pubmed - Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, Solomon D, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis. 2013;17(5 Suppl 1):S1-S27.
doi pubmed - Witt BL, Factor RE, Jarboe EA, Layfield LJ. Negative loop electrosurgical cone biopsy finding following a biopsy diagnosis of high-grade squamous intraepithelial lesion: frequency and clinical significance. Arch Pathol Lab Med. 2012;136(10):1259-1261.
doi pubmed - Ryu A, Nam K, Chung S, Kim J, Lee H, Koh E, Bae D. Absence of dysplasia in the excised cervix by a loop electrosurgical excision procedure in the treatment of cervical intraepithelial neoplasia. J Gynecol Oncol. 2010;21(2):87-92.
doi pubmed - Magkana M, Mentzelopoulou P, Magkana E, Pampanos A, Vrachnis N, Kalafati E, Daskalakis G, et al. p16/Ki-67 dual staining is a reliable biomarker for risk stratification for patients with borderline/mild cytology in cervical cancer screening. Anticancer Res. 2022;42(5):2599-2606.
doi pubmed - Luan T, Hua Q, Liu X, Xu P, Gu Y, Qian H, Yan L, et al. PAX1 methylation as a potential biomarker to predict the progression of cervical intraepithelial neoplasia: a meta-analysis of related studies. Int J Gynecol Cancer. 2017;27(7):1480-1488.
doi pubmed - Muresu N, Puci MV, Sotgiu G, Sechi I, Usai M, Cossu A, Martinelli M, et al. Diagnostic accuracy of DNA-methylation in detection of cervical dysplasia: findings from a population-based screening program. Cancers (Basel). 2024;16(11):1986.
doi pubmed - Bonde J, Floore A, Ejegod D, Vink FJ, Hesselink A, van de Ven PM, Valencak AO, et al. Methylation markers FAM19A4 and miR124-2 as triage strategy for primary human papillomavirus screen positive women: A large European multicenter study. Int J Cancer. 2021;148(2):396-405.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
World Journal of Oncology is published by Elmer Press Inc.