Figures
Figure 1. Flow diagram.
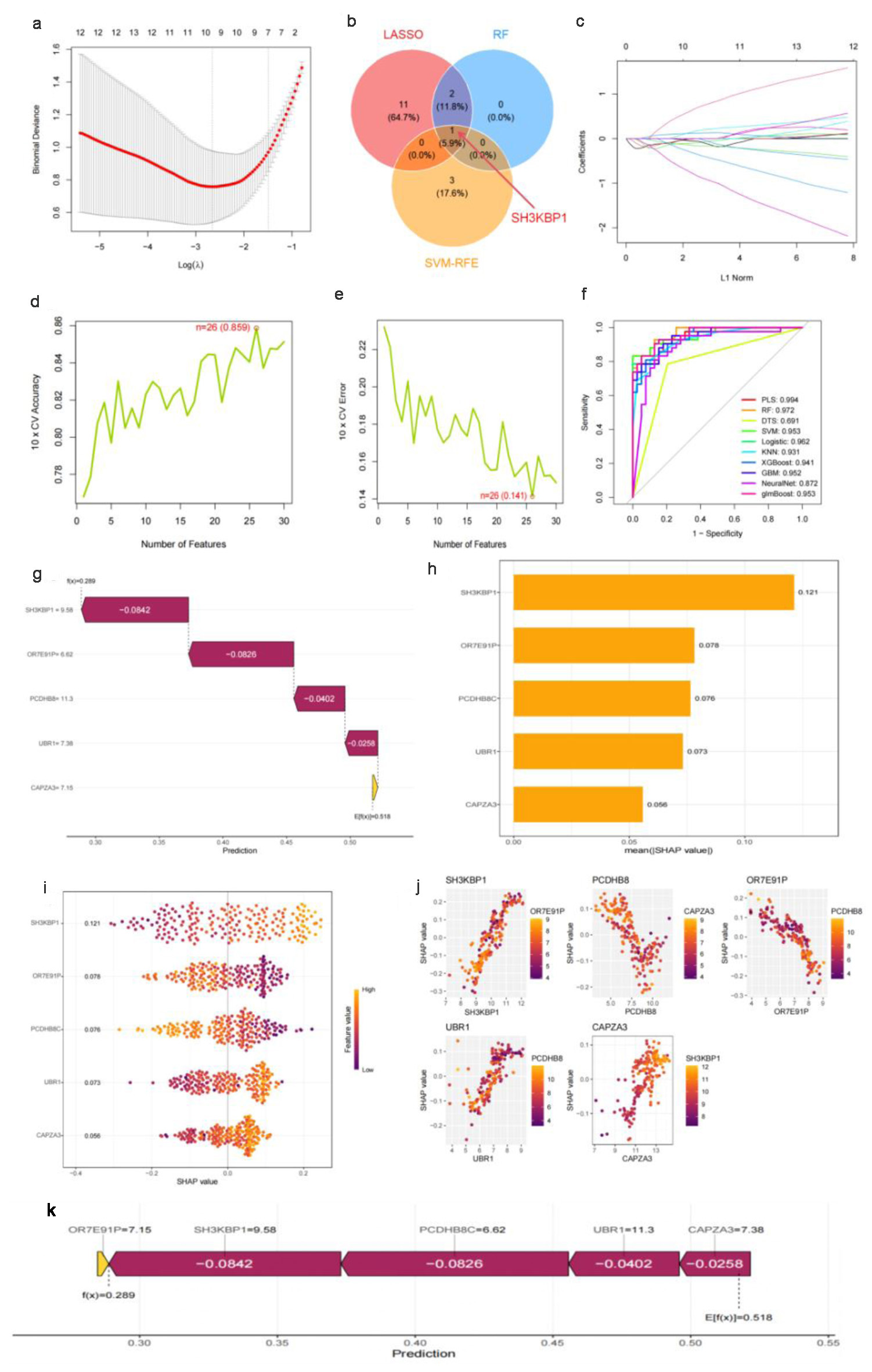
Figure 2. Machine learning feature selection and SHAP interpretability analysis based on multiple datasets. (a) Ten-fold cross-validation λ parameter selection curve for LASSO regression, with the optimal λ value marked by a red vertical line. (b) Venn diagram of feature selection results from three machine learning algorithms (LASSO, RF, SVM-RFE), with SH3KBP1 located in the intersection area of the algorithms. (c) LASSO regression coefficient shrinkage path diagram. (d) Ten-fold cross-validation accuracy curve with the number of features, showing a maximum accuracy of 0.850. (e) Ten-fold cross-validation error rate curve, showing a minimum error rate of 0.141. (f) ROC performance comparison of multiple machine learning algorithms, with the PLS algorithm having the highest AUC (0.994). (g) Univariate prediction efficiency analysis of key genes. (h) SHAP feature importance ranking diagram. (i) SHAP value summary diagram, showing the distribution of the impact of gene expression on prediction. (j) SHAP dependence diagram, showing the interaction between SH3KBP1 and such genes as PCDH8B, OR7E31P, USR1, and CAPZA3. (k) Flowchart of the final prediction model constructed based on key genes.
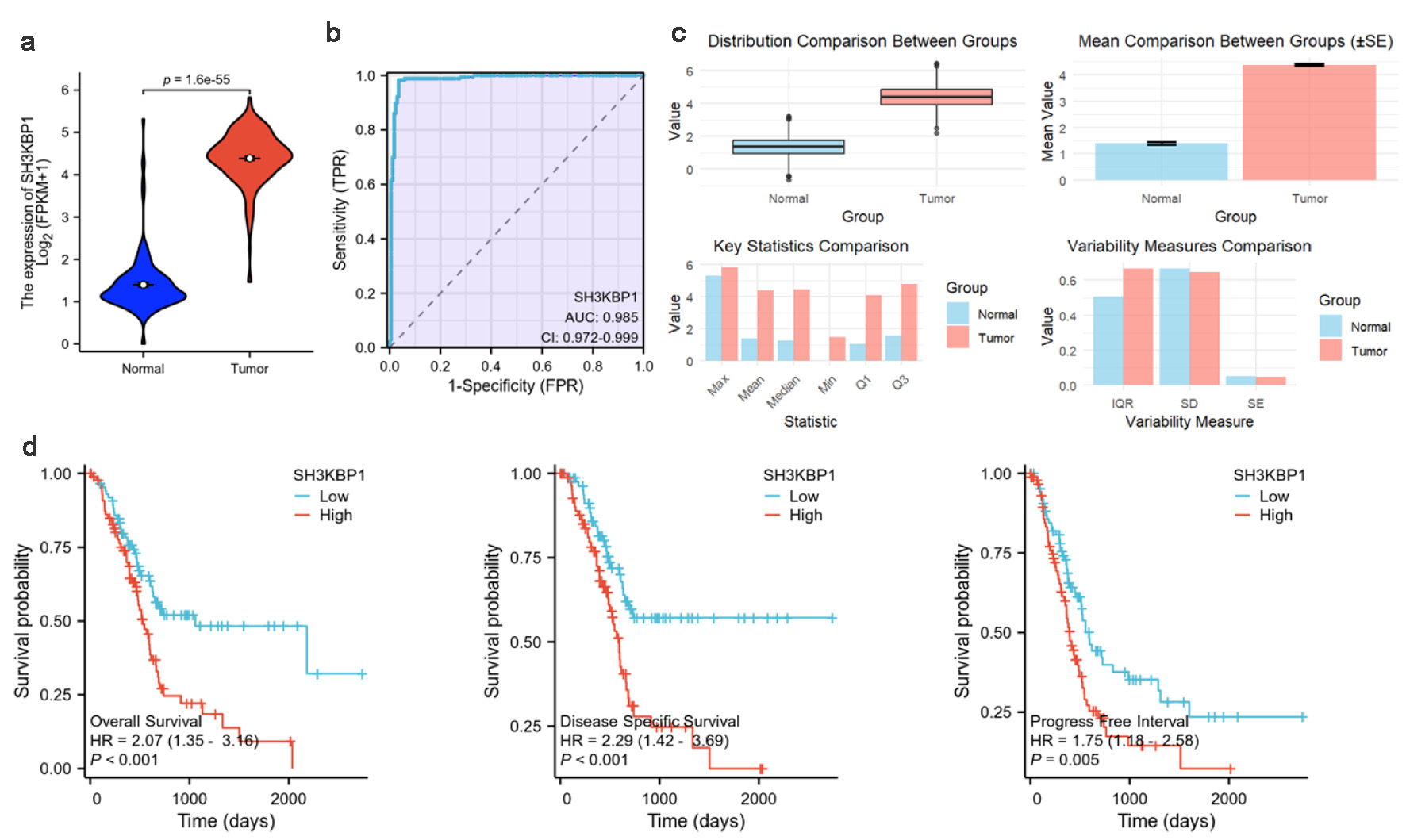
Figure 3. SH3KBP1 expression and its clinical correlation analysis in the TCGA-PDAC dataset. (a) Expression distribution of SH3KBP1 in normal groups and tumor groups. (b) ROC diagnostic curve analysis of SH3KBP1. (c) Comparison of expression level distribution, mean value, and statistical characteristics. (d) Kaplan-Meier survival analysis of SH3KBP1 expression level with OS, DSS, and PFI.
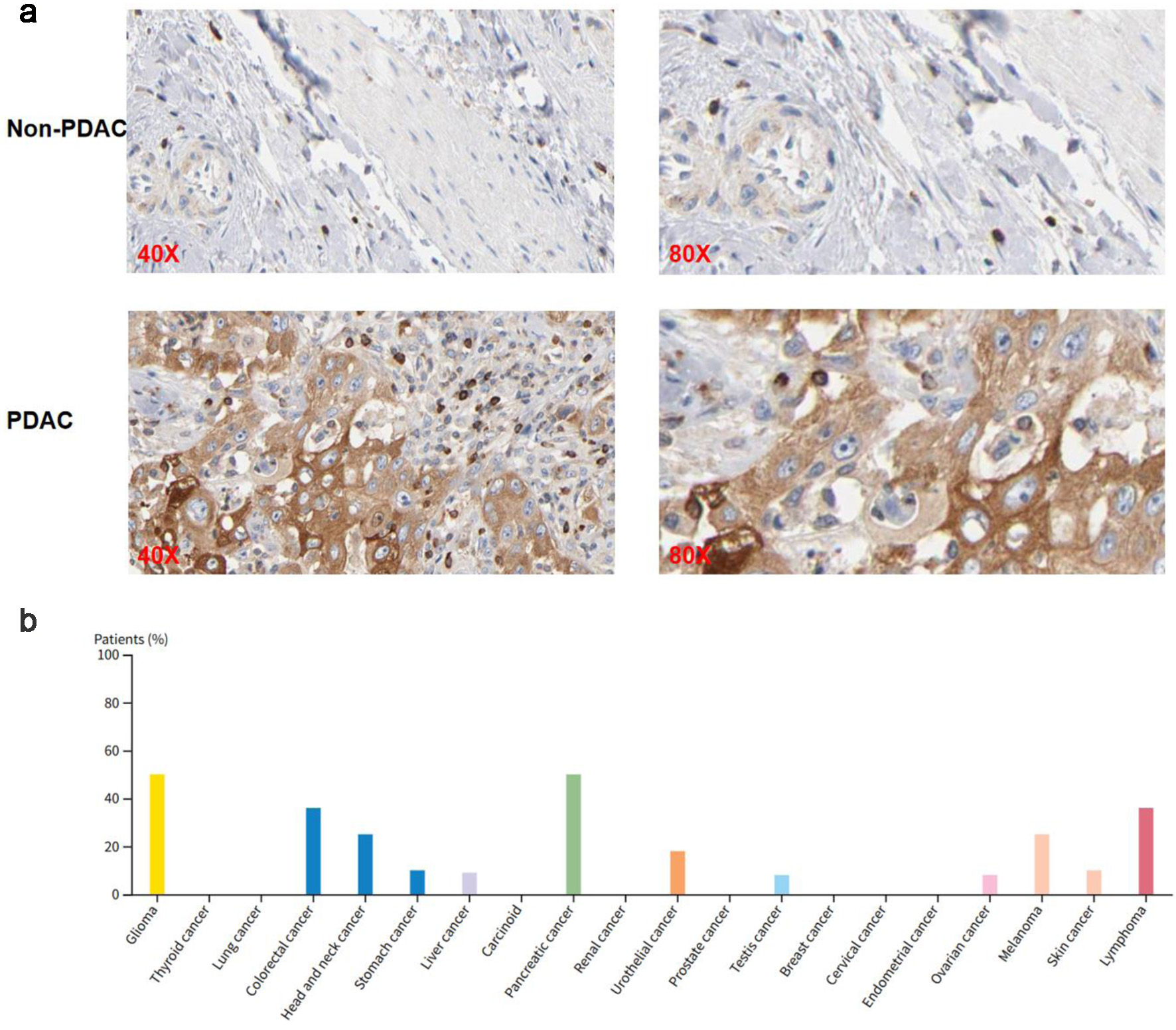
Figure 4. SH3KBP1 protein expression analysis based on the THPA (Antibody: HPA003351). (a) Immunohistochemical images showing the expression of SH3KBP1 in non-PDAC and PDAC tissues, with magnification levels of × 40 and × 80. (b) Distribution of the proportion of positive patients with SH3KBP1 in various tumor types.
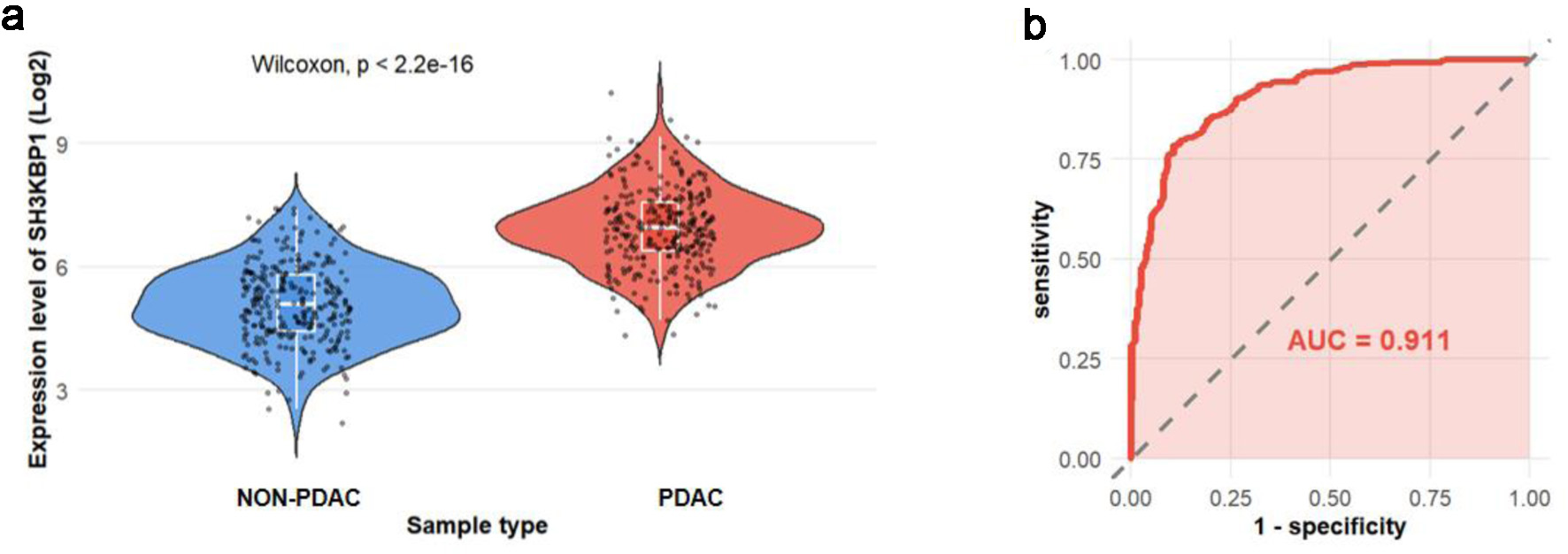
Figure 5. SH3KBP1 protein expression analysis based on the PDC database. (a) Violin plot showing the expression distribution of SH3KBP1 in non-PDAC and PDAC samples (300 cases in each group). (b) ROC curve evaluating the discriminative ability of SH3KBP1 in PDAC (AUC = 0.911).
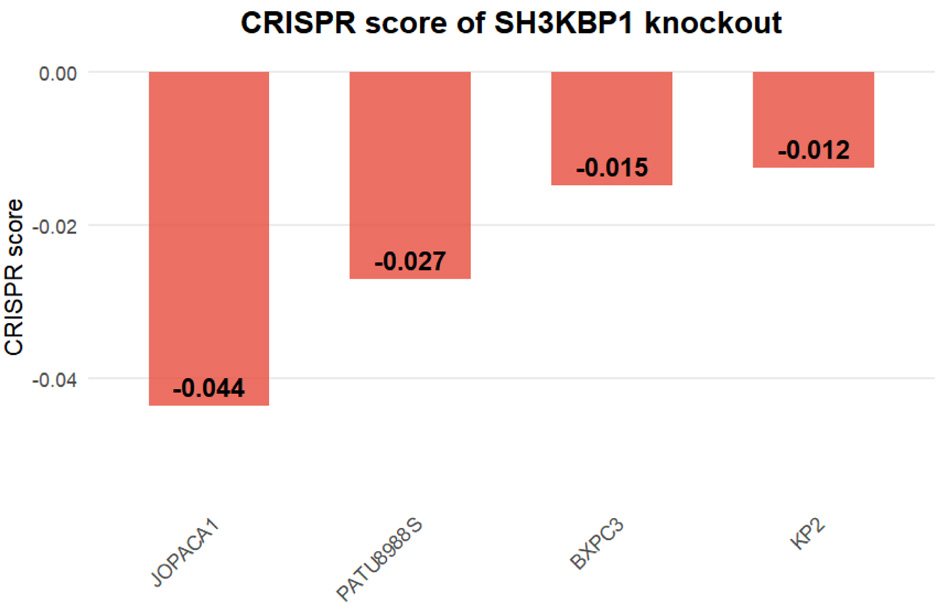
Figure 6. CRISPR knockout score of the SH3KBP1 gene in PDAC cell lines based on the DepMap database. Bar chart showing the CRISPR dependency scores (CRISPR score) of SH3KBP1 in four PDAC cell lines (JOPACA1, PATU8988S, BXPC3, and KP2).
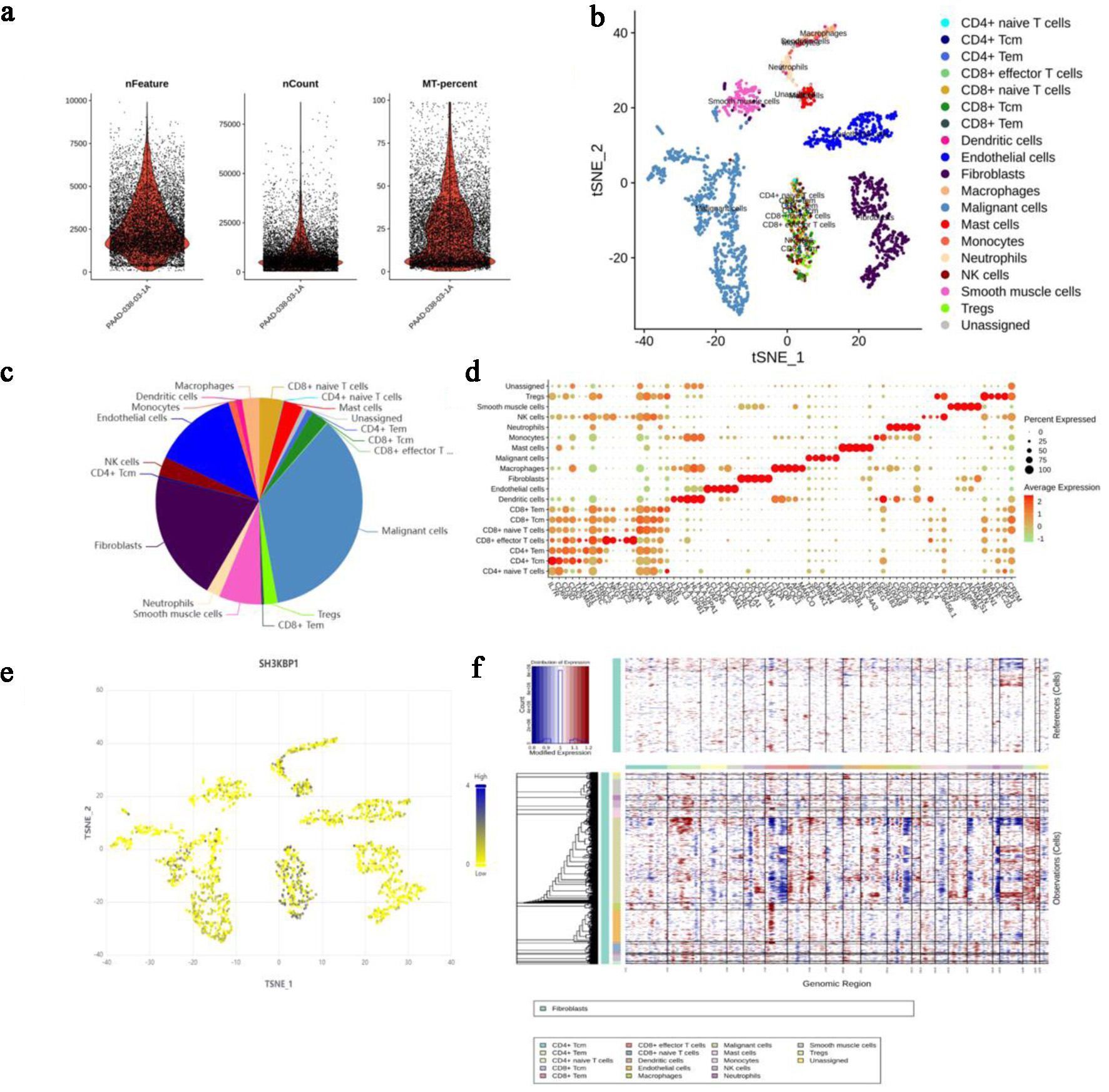
Figure 7. Analysis of cell heterogeneity and SH3KBP1 expression in PDAC tissues based on single-cell sequencing data from the NCBI SRA project (accession number: PRJNA885258). (a) Sample quality control parameters, including the number of genes (nFeature), number of UMIs (nCount), and mitochondrial gene ratio (MT.percent). (b) t-SNE dimensionality reduction map showing the spatial distribution of different cell populations. (c) Pie chart showing the composition ratio of various cell types in the sample. (d) DotPlot showing the expression levels and expression proportions of characteristic marker genes in each cell population. (e) Expression distribution of SH3KBP1 in various cell populations (t-SNE map). (f) Inference of whether cells are malignant based on single-cell CNV analysis, with the upper panel showing reference cells (e.g., T cells) and the lower panel showing the CNV heatmap of other cells.
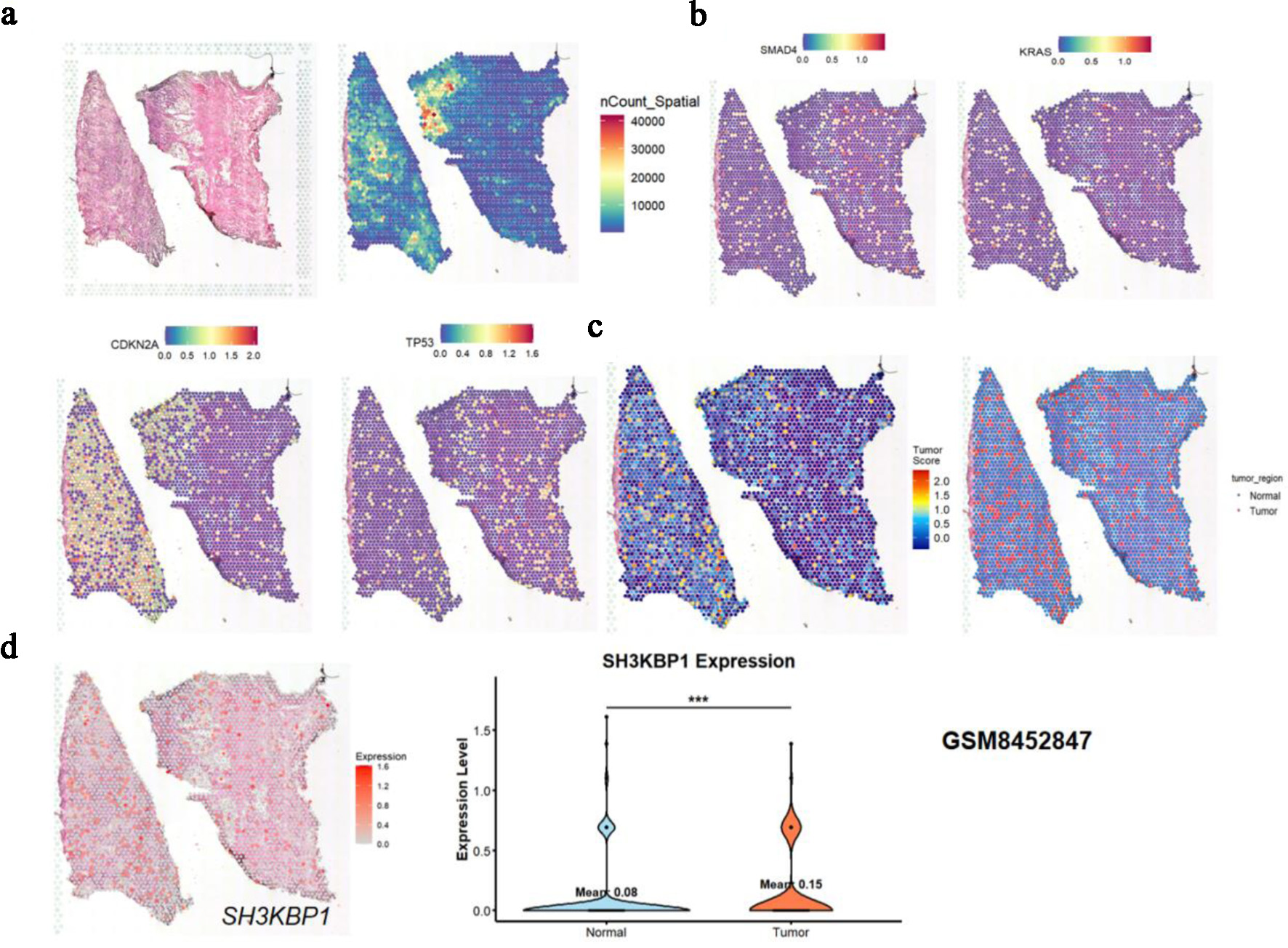
Figure 8. Spatial expression analysis of SH3KBP1 based on GSM8452847 spatial transcriptome data. (a) Spatial tissue structure map, spatial UMI count map (nCount_Spatial), and expression distribution of tumor key genes (CDKN2A, TP53) in tissue sections. (b) Spatial expression distribution of common driver genes (SMAD4, KRAS) in PDAC. (c) Tumor region heatmap and binary classification region map (Tumor vs. Normal) divided according to the expression patterns of key molecules (e.g., KRAS, TP53) in (b). (d) Spatial expression heatmap of SH3KBP1 and violin plot of its expression difference analysis between tumor regions and normal regions.
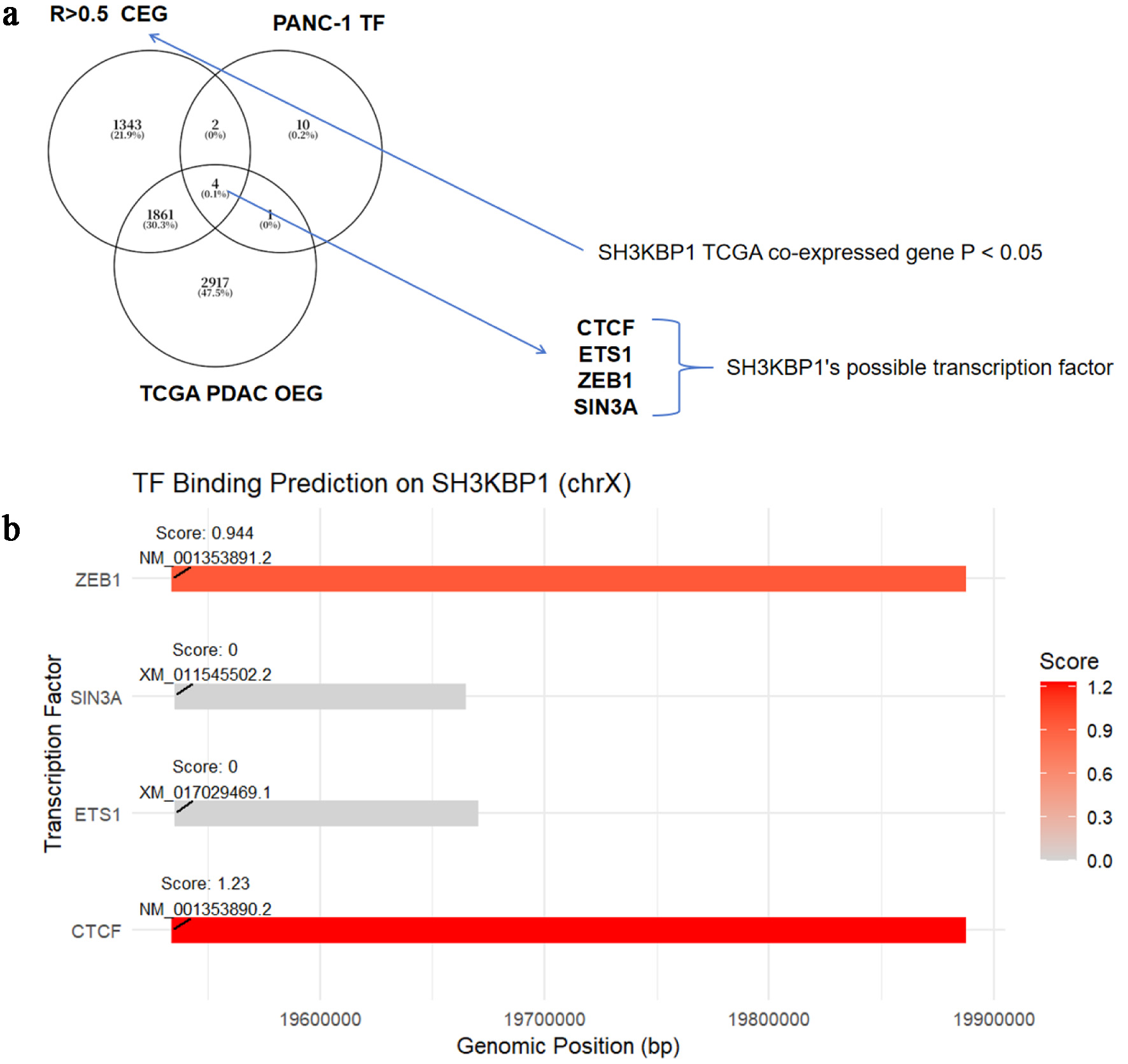
Figure 9. Prediction analysis of potential transcription factors of SH3KBP1 based on the CDB database. (a) Venn diagram showing the intersection of SH3KBP1-coexpressed genes (P < 0.05) in TCGA-PDAC, upregulated DEG, and TF targets in the PANC-1 cell line, screening out four candidate transcription factors (CTCF, ETS1, ZEB1, and SIN3A). (b) Binding sites and scoring of transcription factors predicted by the CDB database in the SH3KBP1 promoter region (chrX).
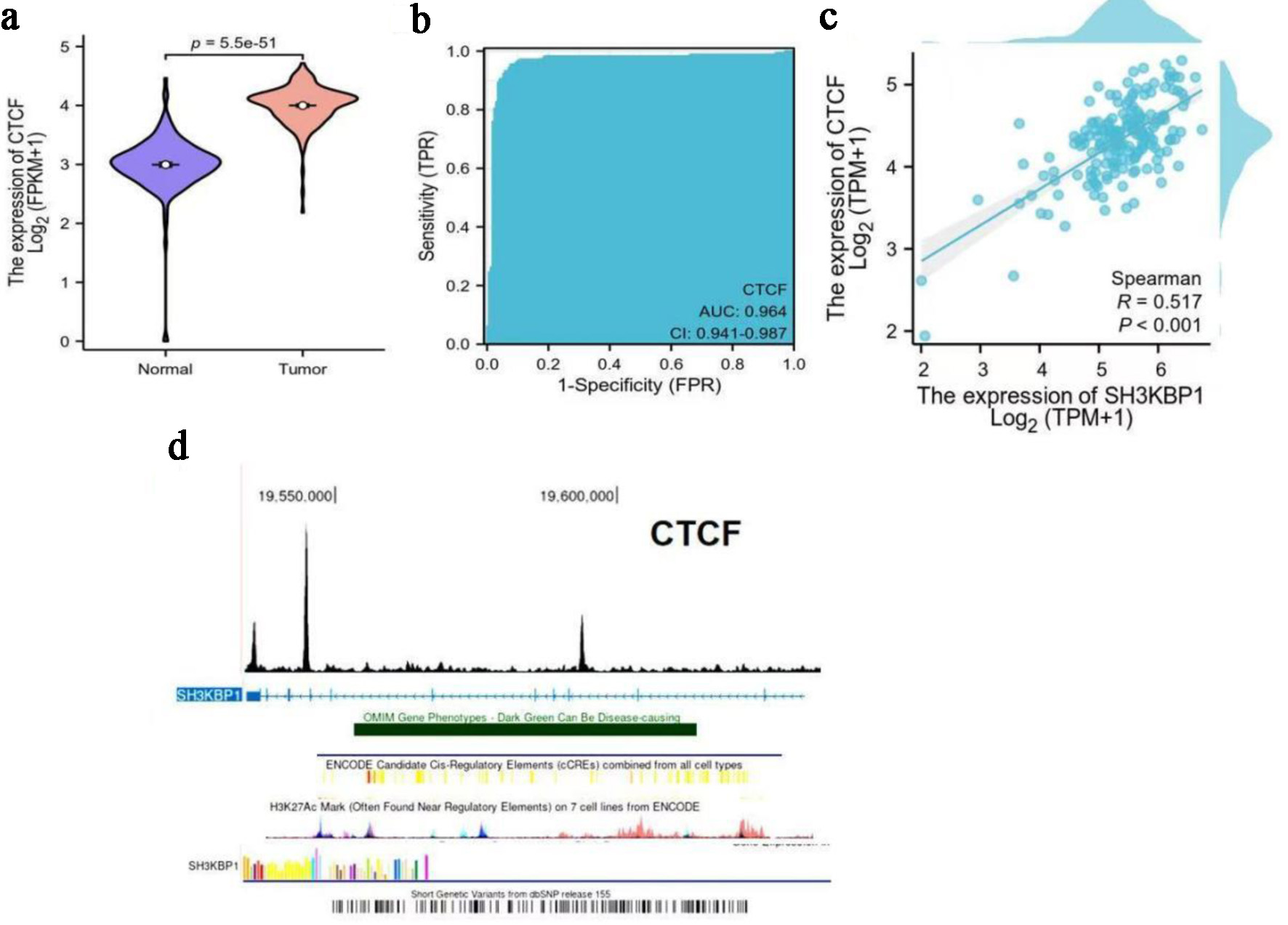
Figure 10. Expression characteristics of CTCF in PDAC and its correlation with SH3KBP1. (a) Analysis of CTCF expression difference between normal tissues and tumor tissues in the TCGA database (Log2(FPKM+1)). (b) ROC curve evaluating the diagnostic efficacy of CTCF for PDAC (AUC). (c) Spearman correlation analysis of SH3KBP1 and CTCF expression levels in PDAC samples (Log2(TPM+1)). (d) ChIP-seq enrichment map of CTCF in the SH3KBP1 gene region (hg38 reference genome), showing its potential binding sites and transcriptional regulatory regions.
Figure 11. Schematic diagram of the role of SH3KBP1 in the PDAC.











