Figures
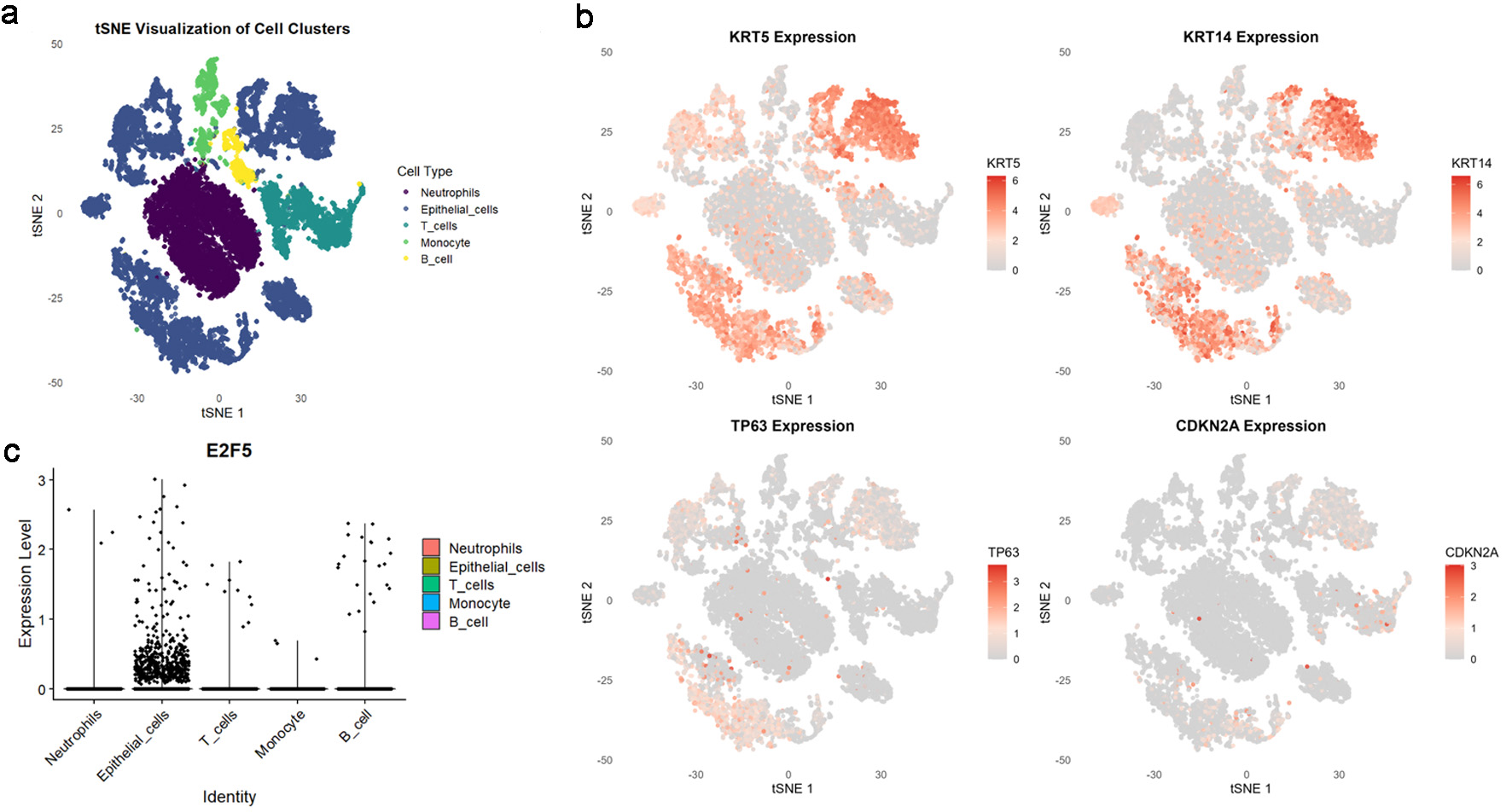
Figure 1. High E2F5 expression in LSCC cells. (a) tSNE plot showing the distribution of different cell types in the single-cell population. (b) LSCC cells labeled with KRT5, KRT14, TP63, and CDKN2A. (c) Distribution of E2F5 expression in single-cell RNA sequencing data from LSCC. LSCC: laryngeal squamous cell carcinoma.

Figure 2. Spatial transcriptomics data confirm high expression of E2F5 in LSCC tissue. Violin plot of spatial counts, showing gene expression distribution in LSCC samples, accompanied by the corresponding spatial heat map. Spatial expression map of E2F5 in LSCC tissue for samples. LSCC: laryngeal squamous cell carcinoma.
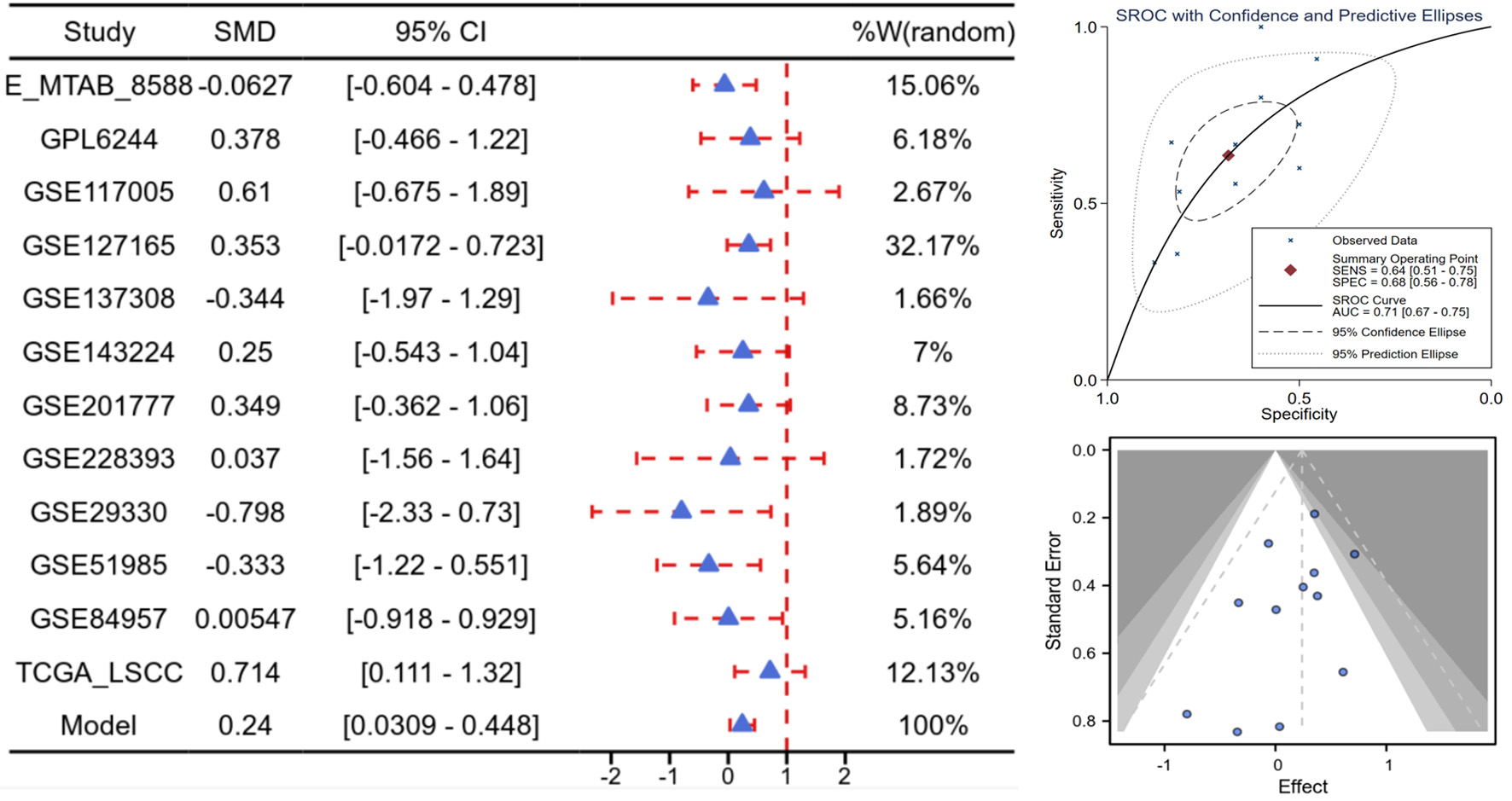
Figure 3. High E2F5 expression in LSCC at the mRNA level. Forest plot of SMD and their 95% CI, showing the means, SD, and SMD for each study’s experimental and control groups. Funnel plot from the Begg test to assess publication bias across studies. sROC curve, presenting the evaluation of discrimination ability. CI: confidence interval; LSCC: laryngeal squamous cell carcinoma; SMD: standardized mean difference; sROC: summary receiver operating characteristic.
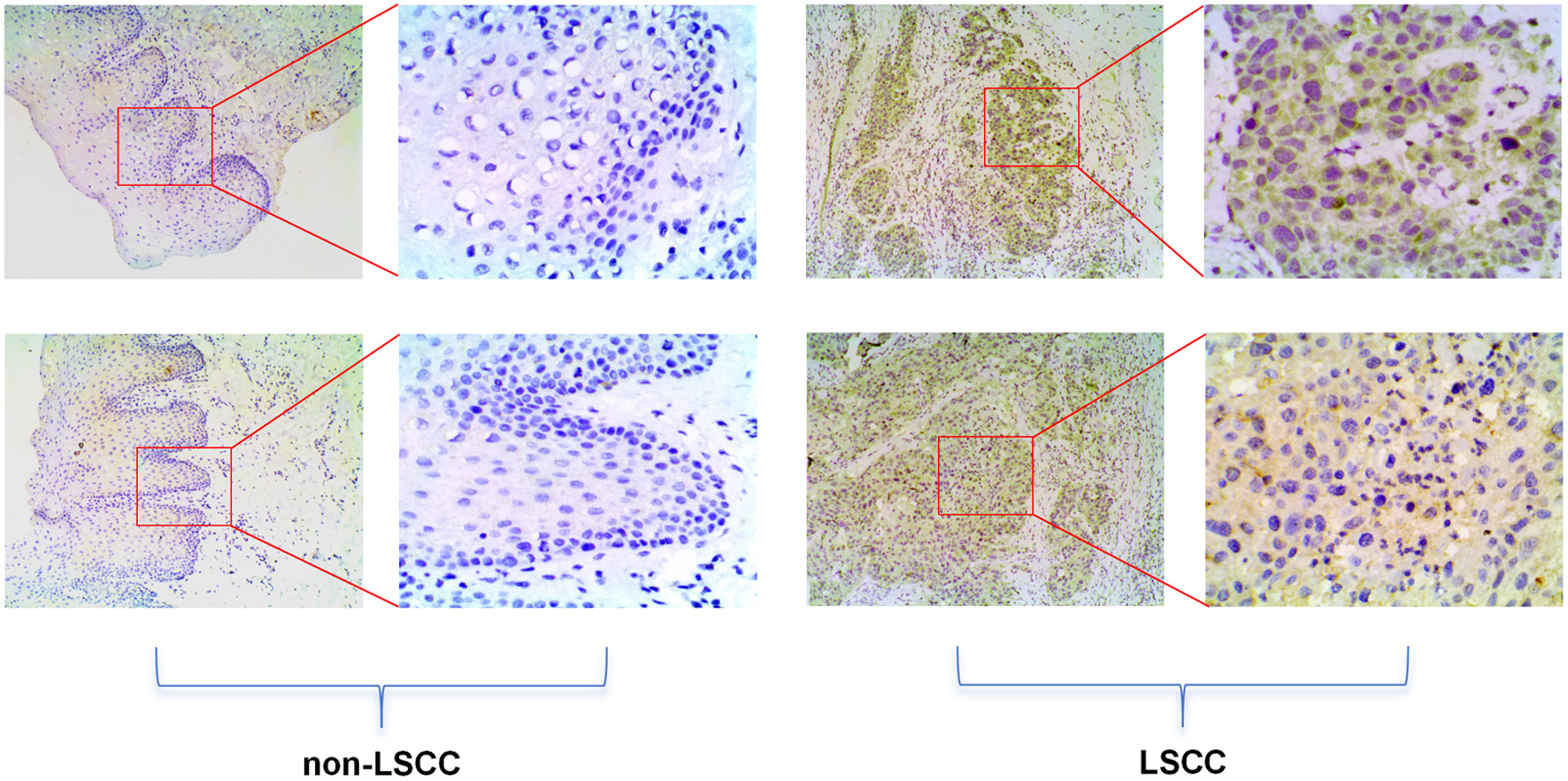
Figure 4. High E2F5 expression in LSCC tissues at the protein level. The top panel shows immunohistochemical staining of non-LSCC tissues, and the bottom panel shows immunohistochemical staining of LSCC tissues. LSCC: laryngeal squamous cell carcinoma.

Figure 5. High E2F5 expression in LSCC tissues at the protein level, as determined by staining counts. (a) Differential expression honeycomb plot. (b) ROC curve. LSCC: laryngeal squamous cell carcinoma; ROC: receiver operating characteristic.
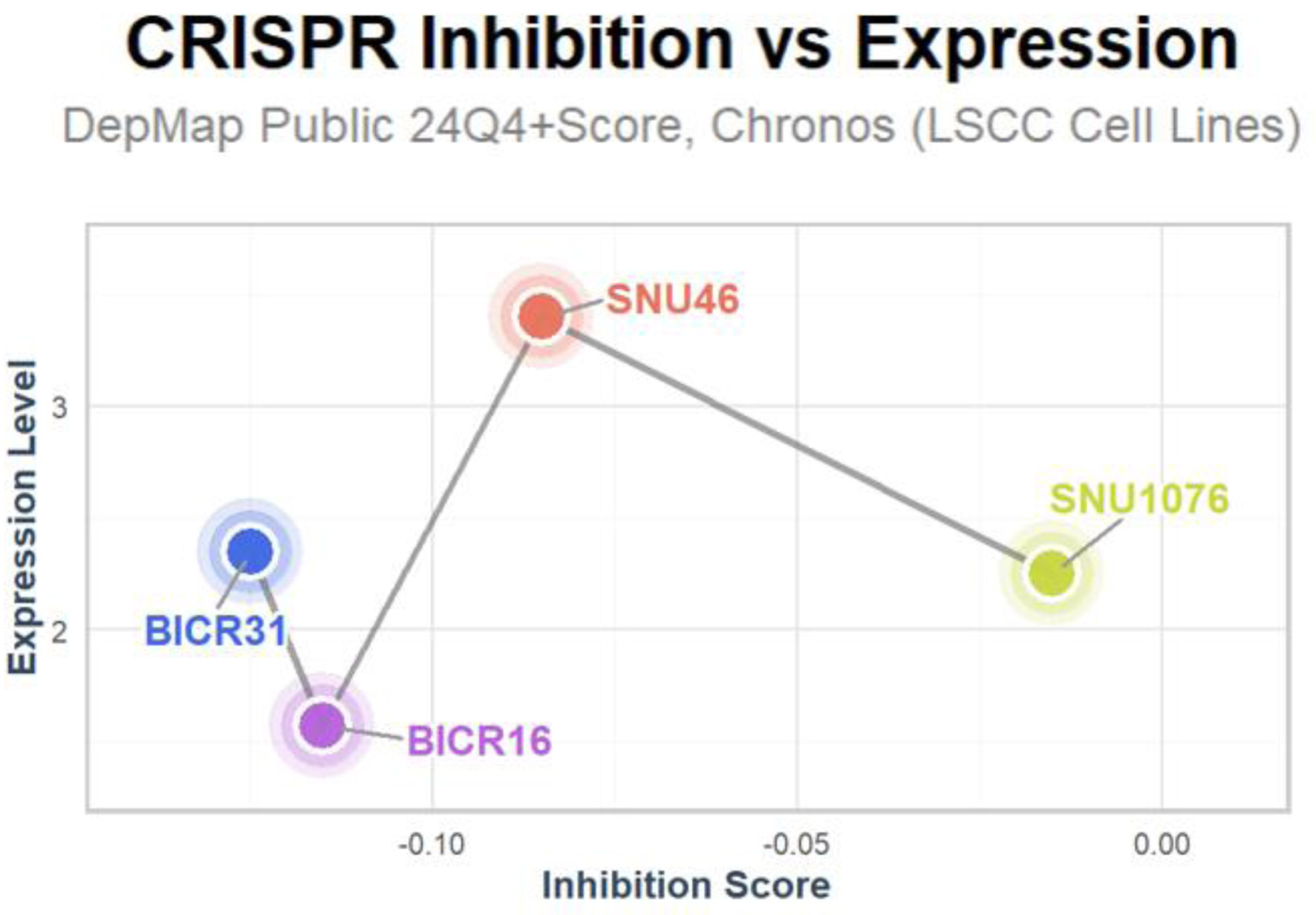
Figure 6. Inhibition of LSCC-related cell lines upon E2F5 knockdown. LSCC: laryngeal squamous cell carcinoma.
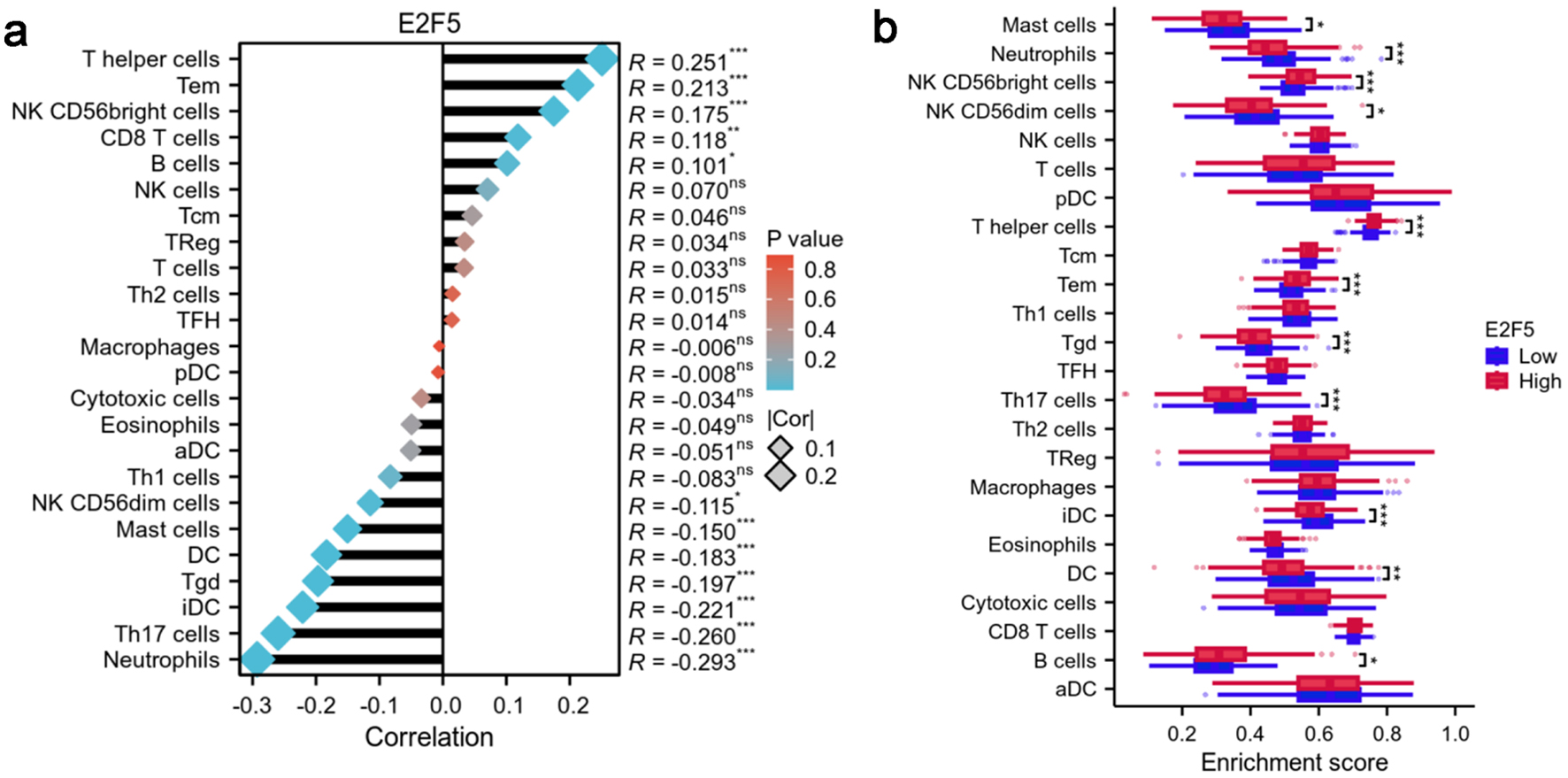
Figure 7. High expression of E2F5 in LSCC affects the enrichment status of immune cells. (a) Correlation between E2F5 expression and immune cells. (b) Differences in immune cells between high and low E2F5 expression groups. Significance symbol: *P < 0.05, **P < 0.01, ***P < 0.001. LSCC: laryngeal squamous cell carcinoma.
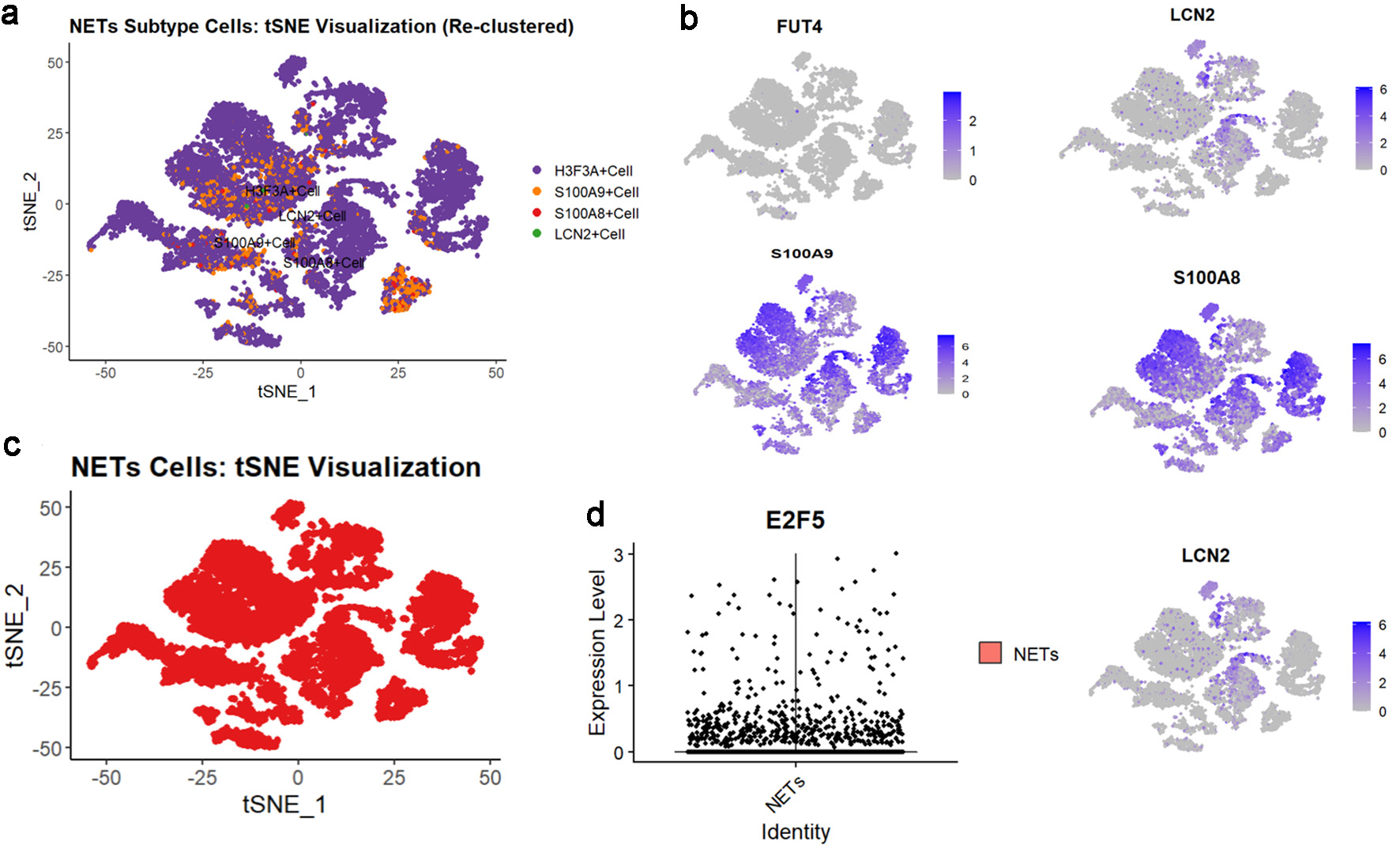
Figure 8. High expression of E2F5 in NETs in LSCC single-cell RNA sequencing. (a) tSNE subtype plot of NETs expression in LSCC. (b) Expression plot of NETs-related indicators. (c) Overall tSNE plot of NETs expression in LSCC. (d) Expression of E2F5 in NETs. LSCC: laryngeal squamous cell carcinoma; NETs: neutrophil extracellular traps.
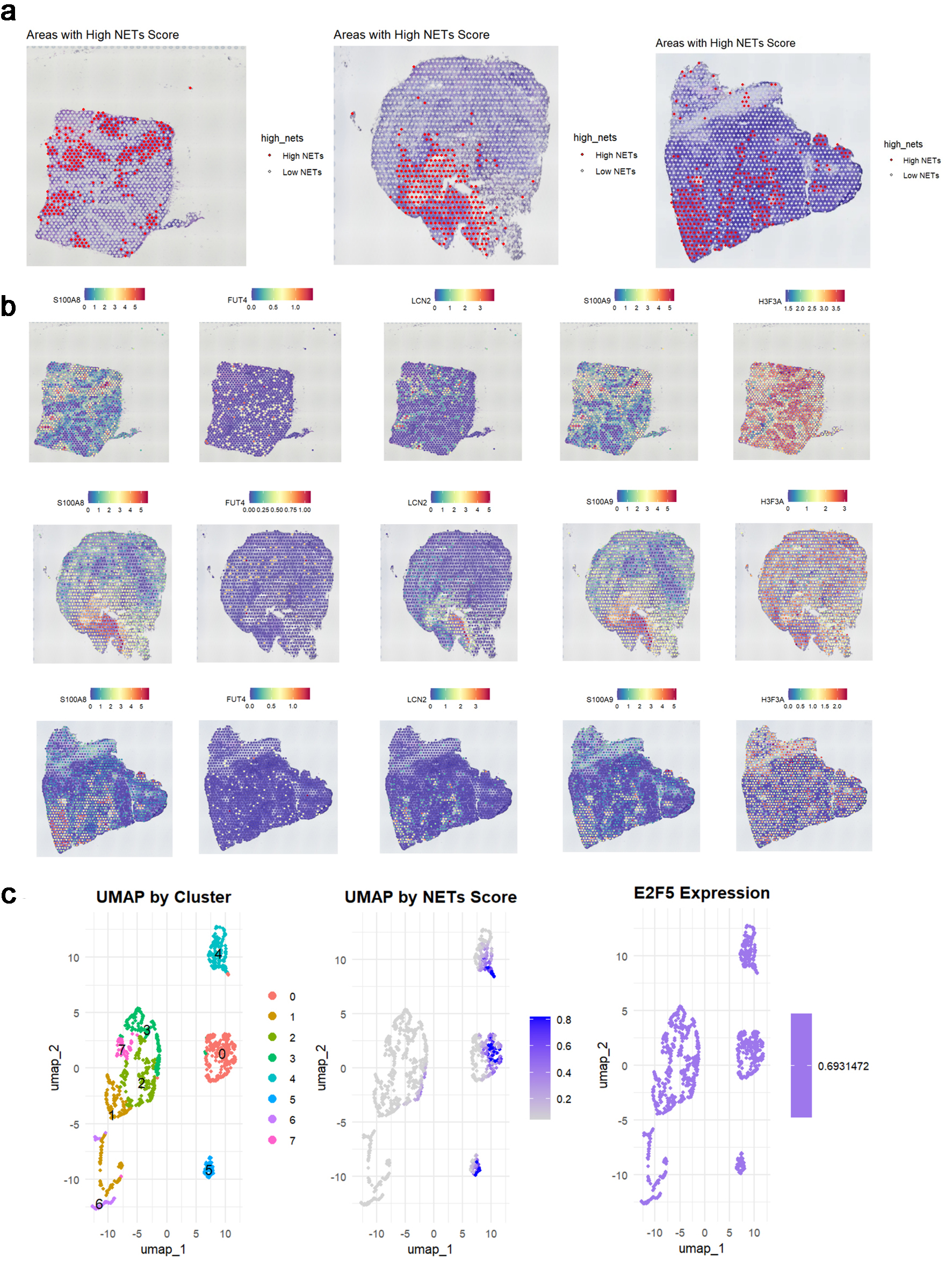
Figure 9. E2F5 expression in LSCC is associated with neutrophil extracellular traps. (a) Spatial distribution of high and low NETs scores across three tumor samples. (b) Spatial expression patterns of key NET-associated markers (S100A8, FUT4, LCN2, S100A9, HSP3A) in corresponding tissue sections. (c) UMAP visualization showing cell clusters, NETs score distribution, and E2F5 expression pattern in single-cell data. LSCC: laryngeal squamous cell carcinoma; NETs: neutrophil extracellular traps.
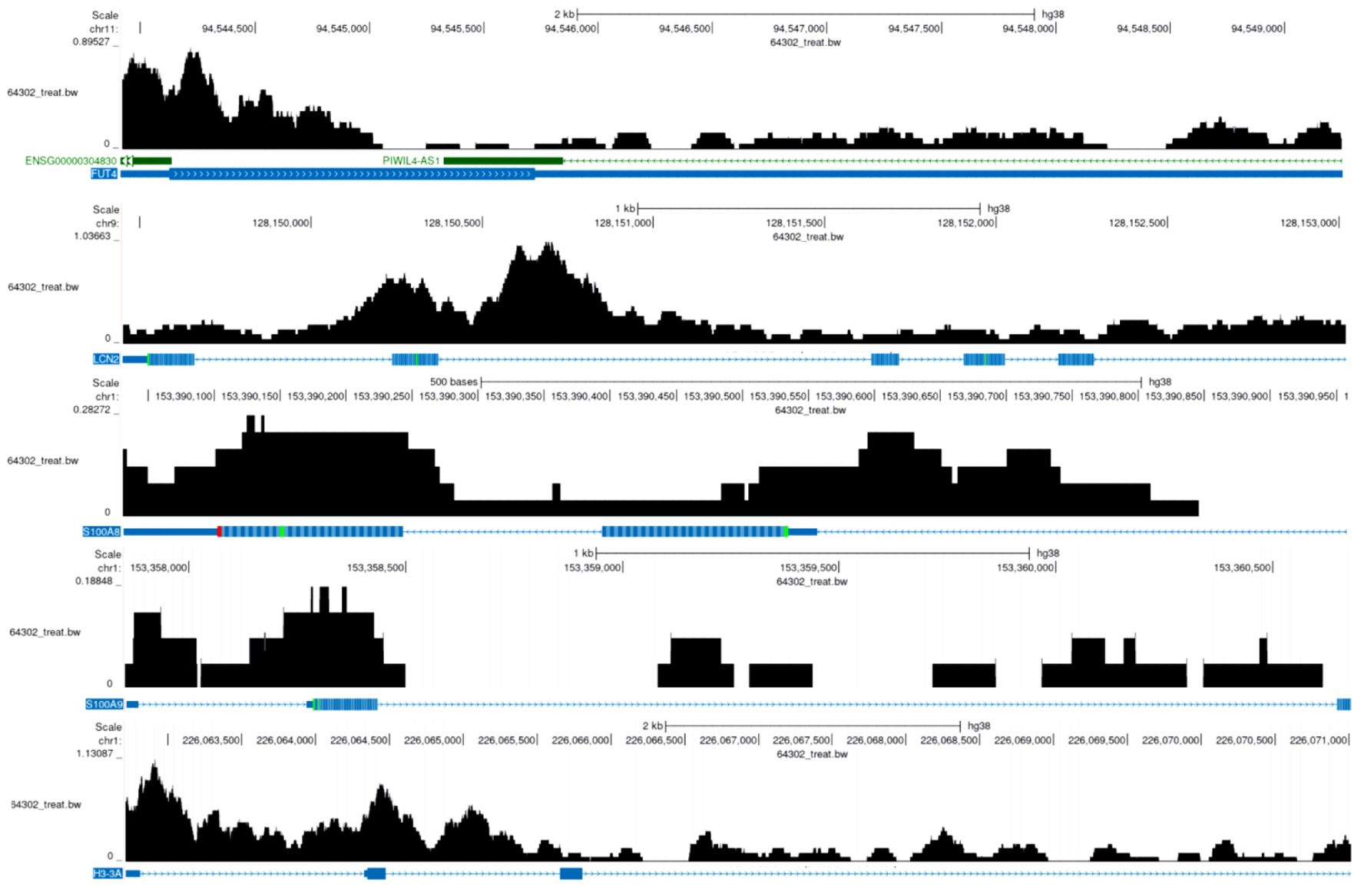
Figure 10. E2F5 transcription factor in NETs-related regions. The black histogram above represents sequencing read depth (64302_treat_bw), the middle colored bands indicate predicted functional elements, and the blue markers below show RefSeq predicted gene models. The gene regions of FUT4, LCN2, S100A8, S100A9, and H3-3A display multiple binding peaks. NETs: neutrophil extracellular traps.
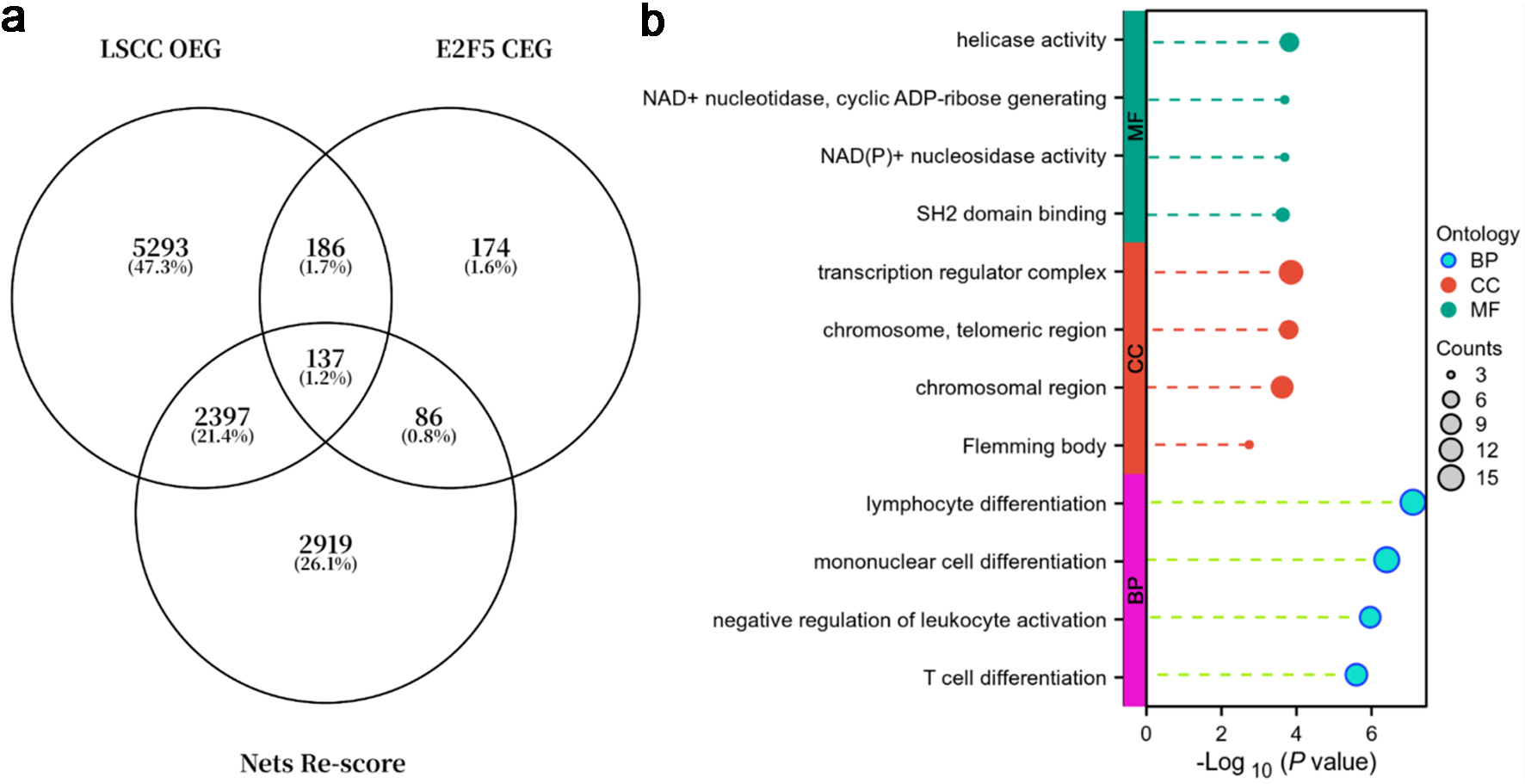
Figure 11. E2F5 and NETs-related genes are primarily enriched in immune regulation and chromosome/transcription regulation. (a) Venn diagram of LSCC overexpressed genes (OEGs), E2F5 co-expressed genes (CEGs), and genes with NETs-related scores > 1.5. (b) Gene function enrichment analysis. LSCC: laryngeal squamous cell carcinoma; NETs: neutrophil extracellular traps.










