Figures
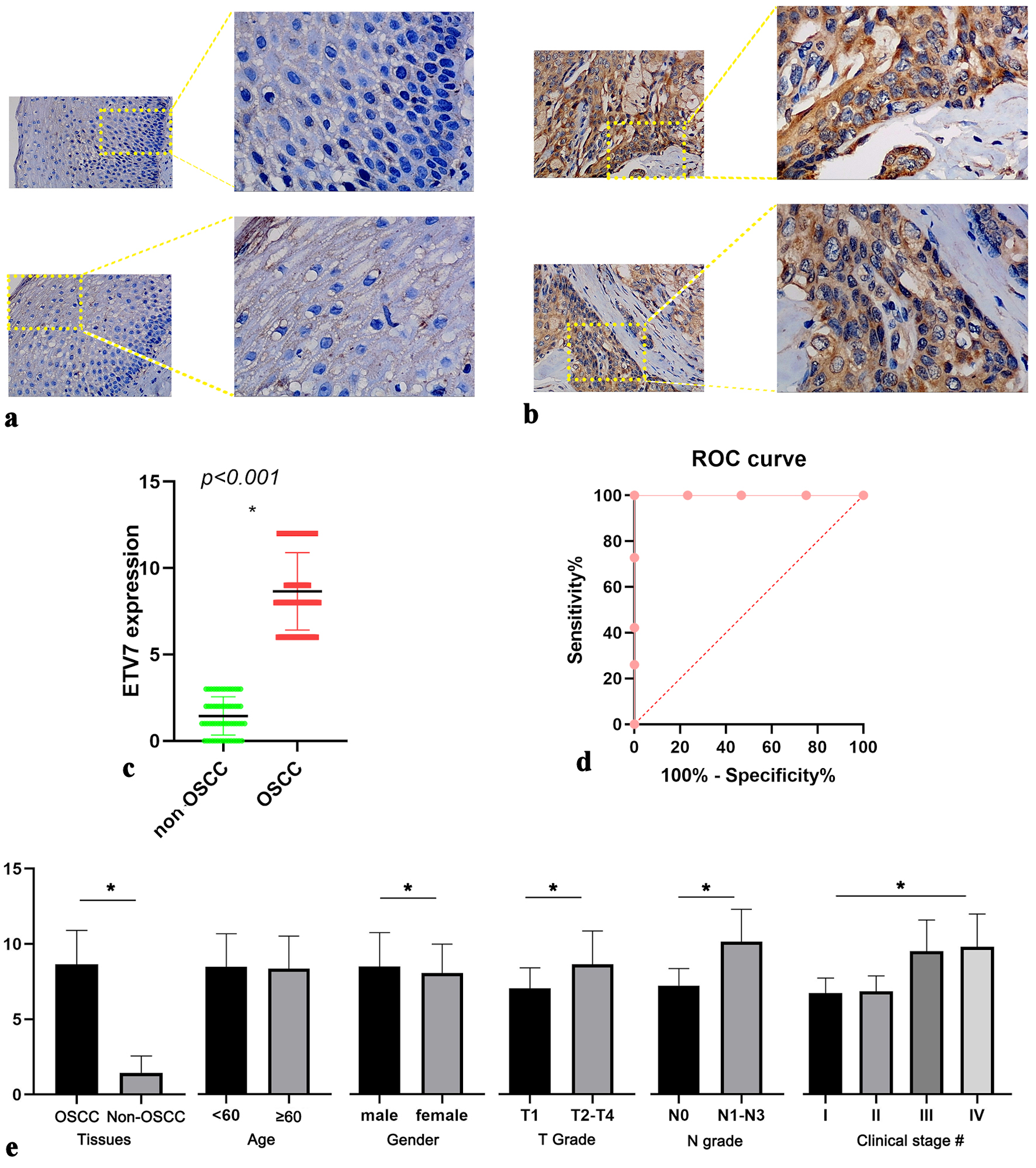
Figure 1. IHC staining of ETV7 expression in OSCC. (a) Expression of ETV7 in non-OSCC tissues. (b) Expression of ETV7 in OSCC tissues. (c) Different expression of ETV7 protein between non-OSCC and OSCC tissues. (d) ROC analysis of ETV7 protein expression in OSCC tissues. (e) Relationship between ETV7 protein level and clinicopathological parameters of OSCC patients based on the IHC. *P < 0.05. #Clinical stage: I vs. III, I vs. IV, II vs. III, II vs. IV, P < 0.001. Early stage (I and II) vs. advanced stage (III and IV) P < 0.001. IHC: immunohistochemistry; ETV7: E26 transformation-specific variant transcription factor 7; OSCC: oral squamous cell carcinoma; ROC: receiver operating characteristic.
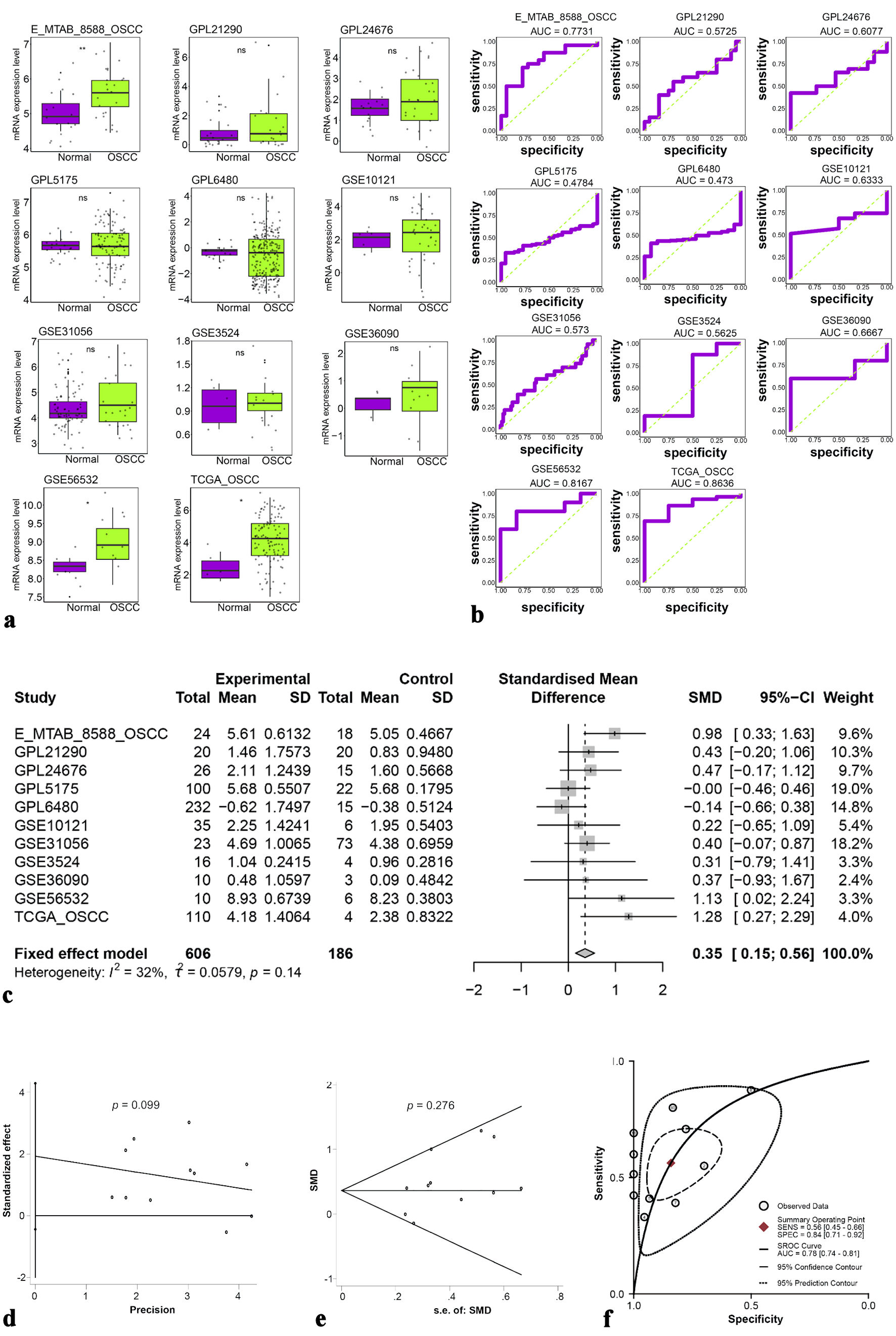
Figure 2. (a) Different expressions of ETV7 mRNA. (b) ROC analysis of ETV7 mRNA expression in each dataset. (c) Forest plot of standardized mean differences of ETV7 mRNA. (d, e) Publication bias test of included datasets. (f) sROC curve of ETV7 mRNA. ETV7: E26 transformation-specific variant transcription factor 7; ROC: receiver operating characteristic; sROC: summary receiver operating characteristic; SD: standard deviation; SMD: standardized mean difference; CI: confidence interval.
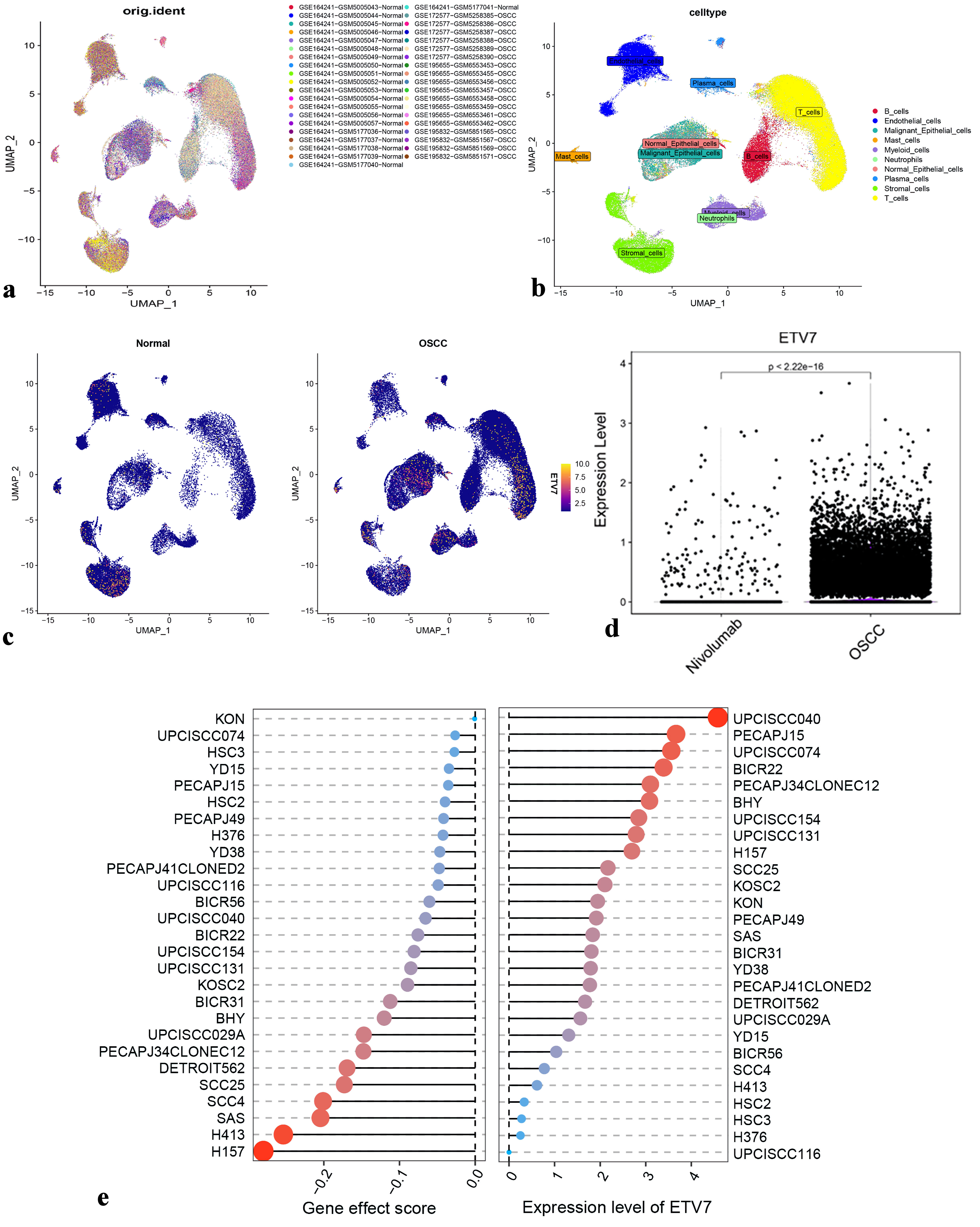
Figure 3. Expression of ETV7 in OSCC different subpopulations. (a) Cluster analysis for scRNA-seq data. (b) Cell annotation for clusters. (c) Density plot of ETV7 expression distribution in each cluster. (c) Changes of OSCC cell subsets after nivolumab treatment. (e) ETV7 expression vs. gene effect in head and neck cancer cell lines. ETV7: E26 transformation-specific variant transcription factor 7; OSCC: oral squamous cell carcinoma; scRNA-seq: single-cell RNA sequencing.
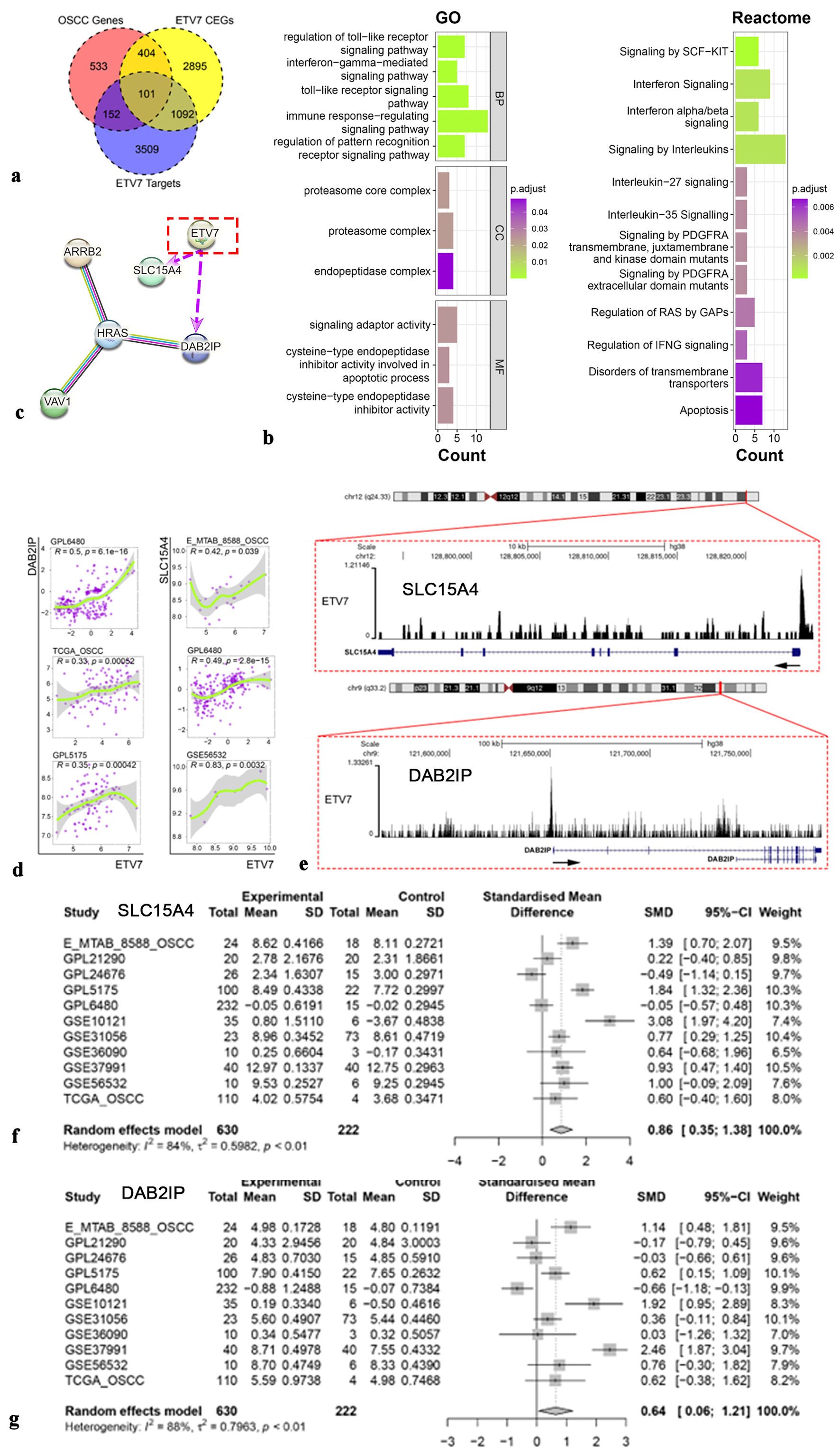
Figure 4. Potential mechanism of ETV7 in OSCC. (a) The intersection of OSCC overexpressed genes, ETV7 coexpressed genes, and ETV7 targets genes. (b) Enrichment analysis for ETV7. (c) Protein-protein interactive network for ETV7 (purple arrows indicate potential positive transcriptional regulatory relationships for ETV7). (d) The relationship between ETV7 and targeted genes SLC15A4 and DAB2IP. (e) SLC15A4 and DAB2IP were potential targets of ETV7. (f) Different expression of SLC15A4 mRNA in OSCC. (g) Different expression of DAB2IP mRNA in OSCC. ETV7: E26 transformation-specific variant transcription factor 7; OSCC: oral squamous cell carcinoma; SD: standard deviation; SMD: standardized mean difference; CI: confidence interval.
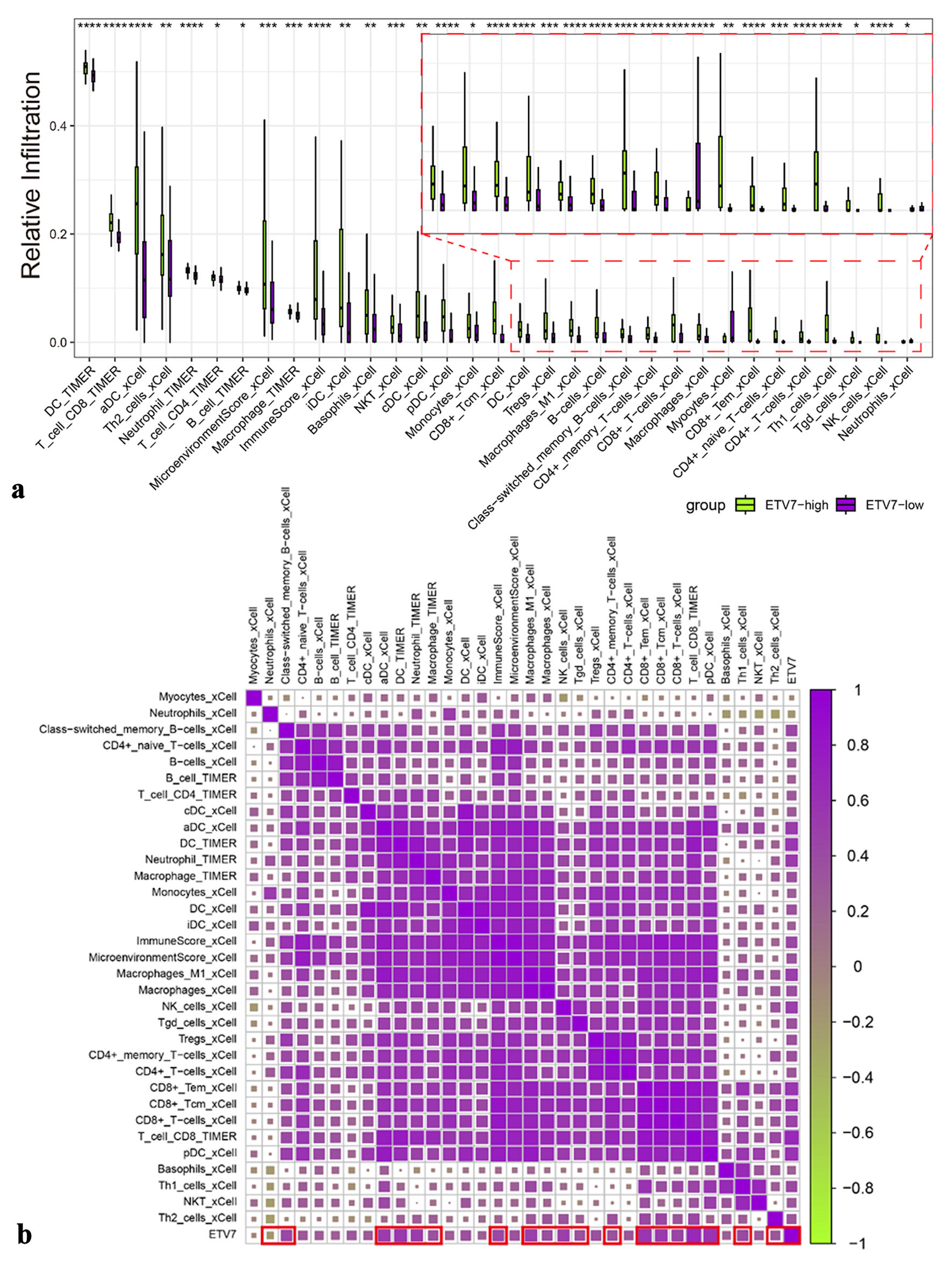
Figure 5. The relationship between immune infiltration and ETV7 expression. (a) Immune infiltration with different expressions of ETV7. (b) Correlation between ETV7 and immune infiltration. ETV7: E26 transformation-specific variant transcription factor 7.
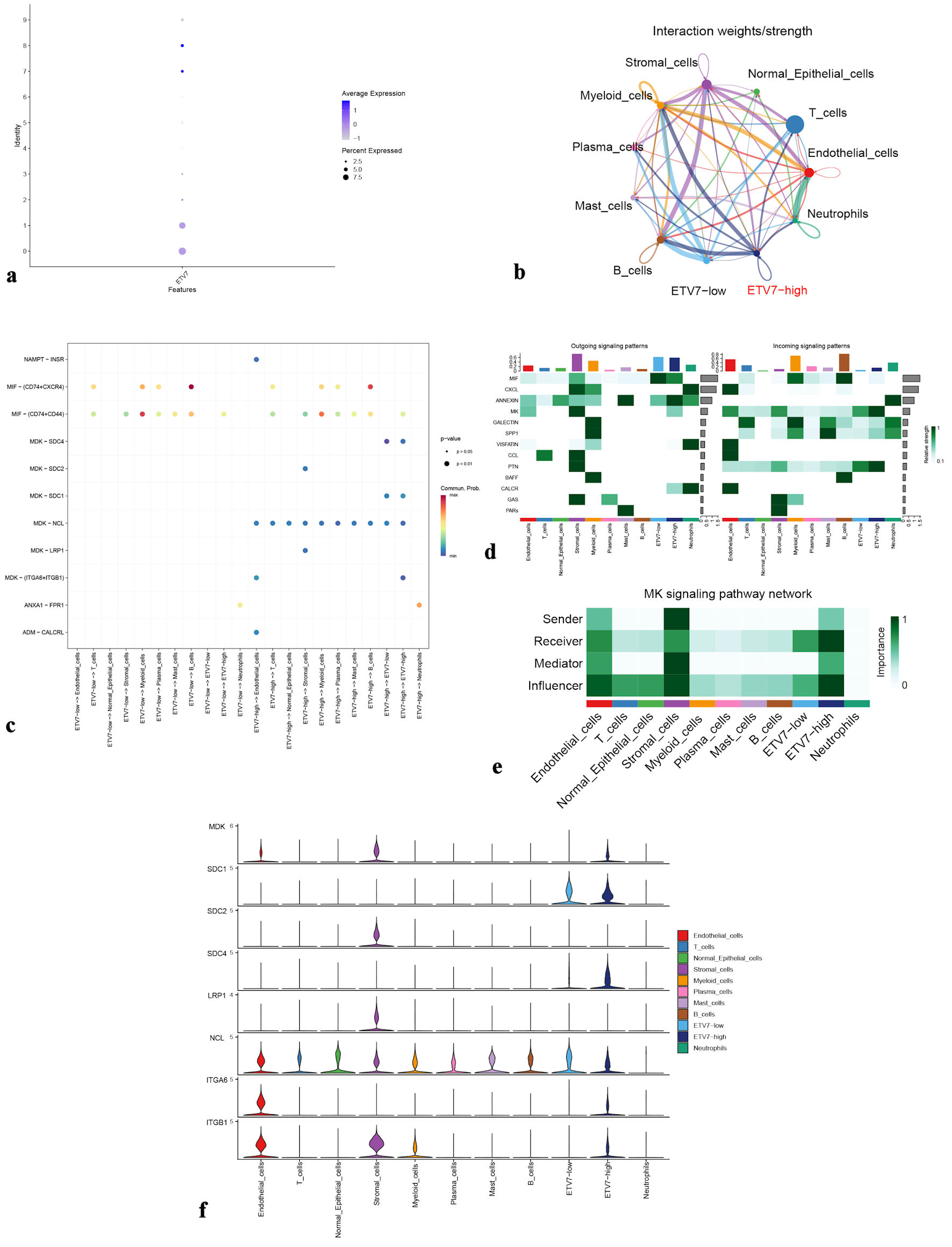
Figure 6. (a) Dot plot showing ETV7 expression in epithelial cell clusters from the OSCC group. (b) Cellular interaction network among different cell types. (c) Ligand-receptor signaling patterns between ETV7 high-expression and low-expression groups. (d) Heatmap of output and input signaling patterns. (e) Midkine (MK) signaling pathway network. (f) Violin plots illustrate expression levels of MK pathway-associated genes. ETV7: E26 transformation-specific variant transcription factor 7; OSCC: oral squamous cell carcinoma.
Table
Table 1. Information of Included High Throughput Data
| Platform | ID | OSCC samples | Non-OSCC samples |
|---|
| OSCC: oral squamous cell carcinoma. |
| GPL24676 | GSE184616 | 15 | 15 |
| GSE179730 | 11 | 0 |
| GPL21290 | GSE186775 | 17 | 17 |
| GSE140707 | 3 | 3 |
| GPL5175 | GSE25099 | 57 | 22 |
| GSE41116 | 43 | 0 |
| GPL6480 | GSE23558 | 27 | 4 |
| GSE46802 | 6 | 10 |
| GSE84846 | 99 | 0 |
| GSE85446 | 66 | 0 |
| GSE85195 | 34 | 1 |
| GPL6353 | GSE10121 | 35 | 6 |
| GPL10526 | GSE31056 | 23 | 73 |
| GPL96 | GSE3524 | 16 | 4 |
| GPL1426 | GSE36090 | 10 | 3 |
| GPL2986 | | | |
| GPL10739 | GSE56532 | 10 | 6 |
| TCGA | OSCC | 110 | 4 |
| E-MTAB | 8588 | 24 | 28 |





