Figures
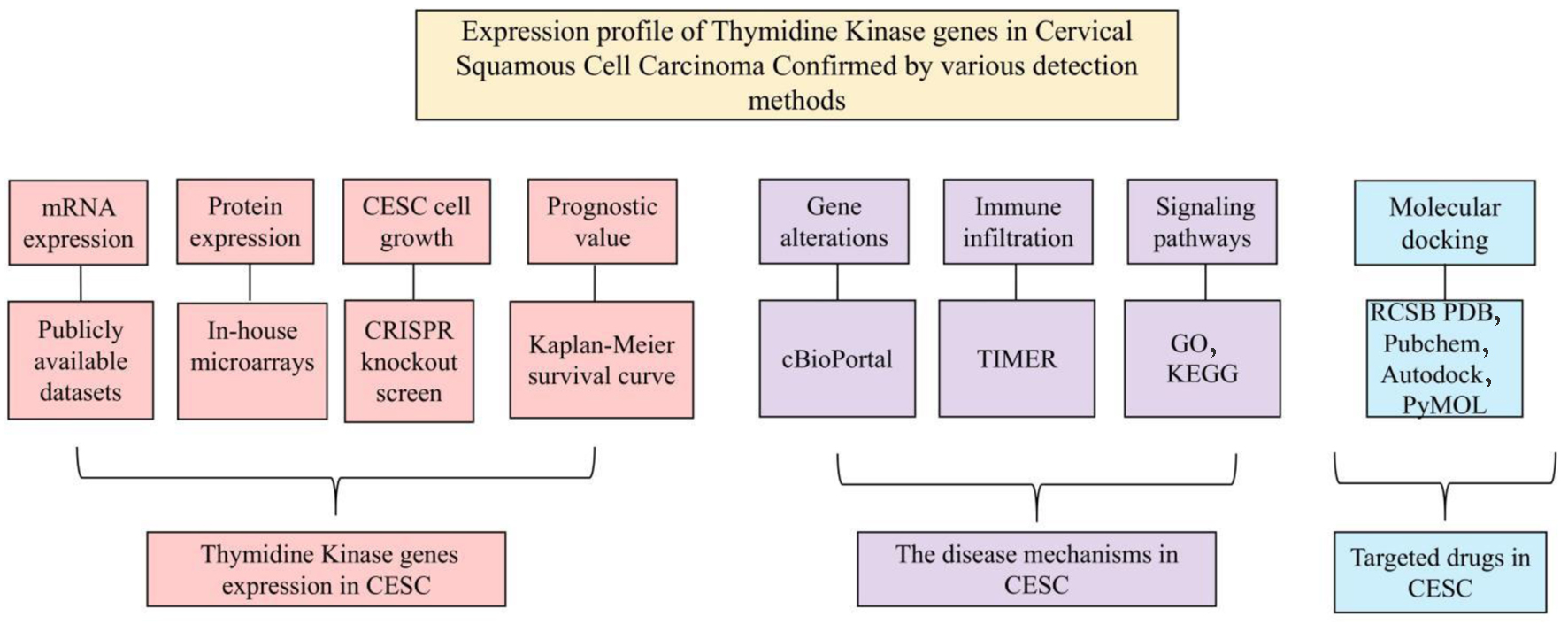
Figure 1. The overall design of the current study.
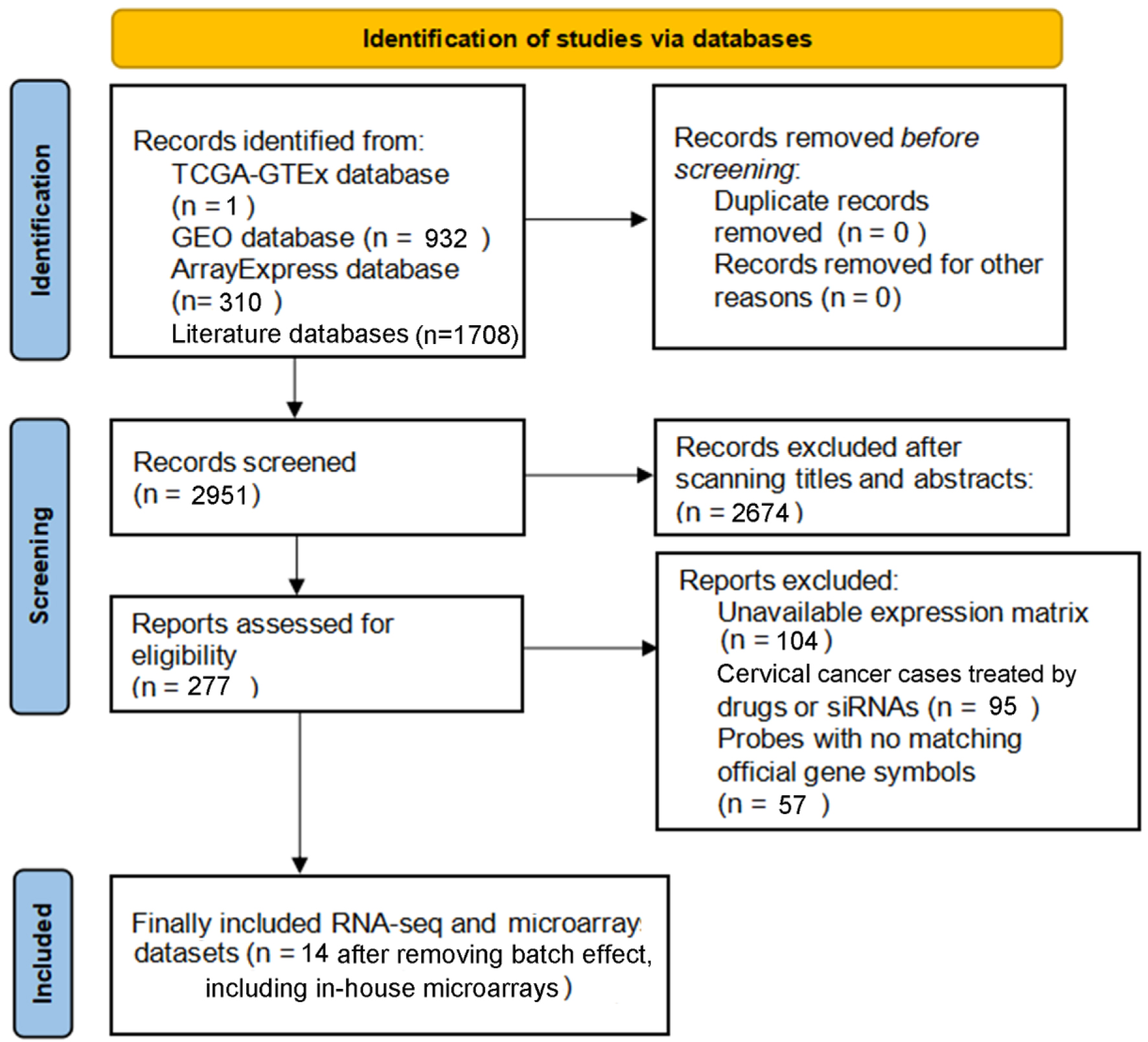
Figure 2. PRISMA flow diagram for the current study.
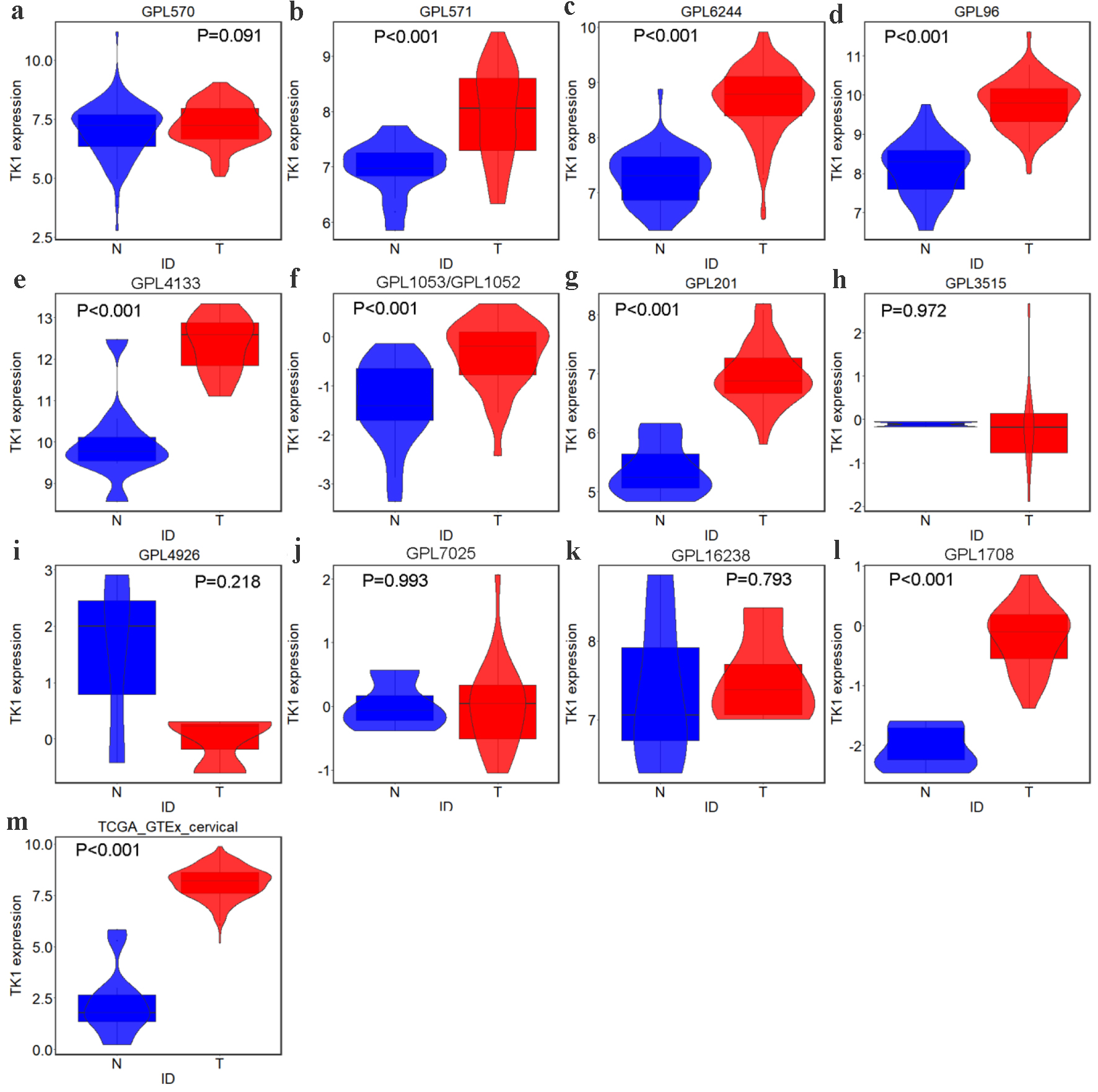
Figure 3. TK1 expression in CESC from external microarrays and RNA-seq datasets. Violin plots for: (a) GPL570; (b) GPL571; (c) GPL6244; (d) GPL96; (e) GPL4133; (f) GPL1053 and GPL1052; (g) GPL201; (h) GPL3515; (i) GPL4926; (j) GPL7025; (k) GPL16238; (l) GPL1708; (m) TCGA-GTEx. N: non-cancer controls; T: CESC samples.
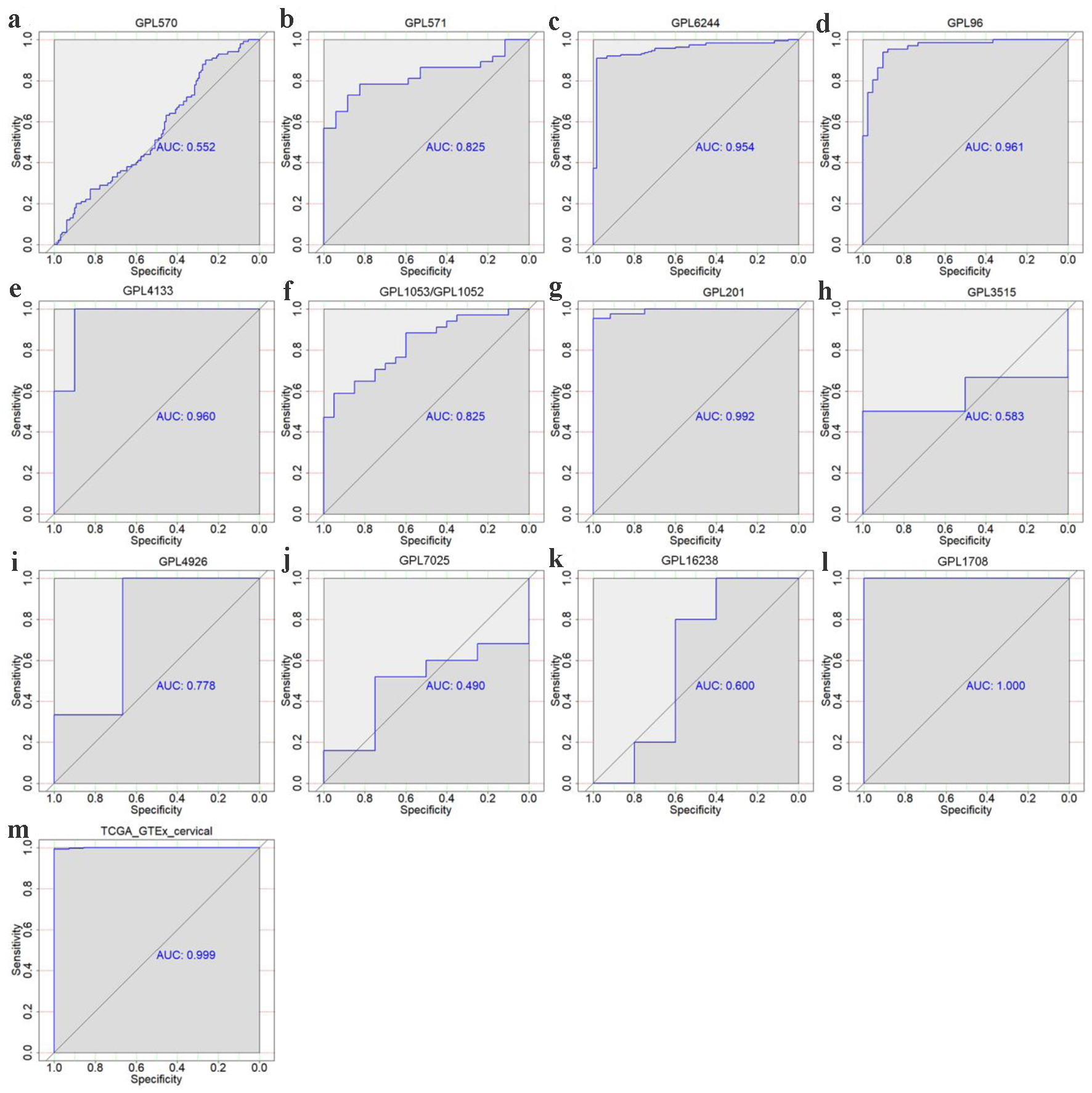
Figure 4. The discriminatory ability of TK1 expression in distinguishing CESC from non-cancer tissues in each microarray and RNA-seq dataset. ROC curves for GPL570 (a), GPL571 (b), GPL6244 (c), GPL96 (d), GPL4133 (e), GPL1053 and GPL1052 (f), GPL201 (g), GPL3515 (h), GPL4926 (i), GPL7025 (j) GPL16238 (k), GPL1708 (l) and TCGA-GTEx datasets (m). AUC: area under curve.
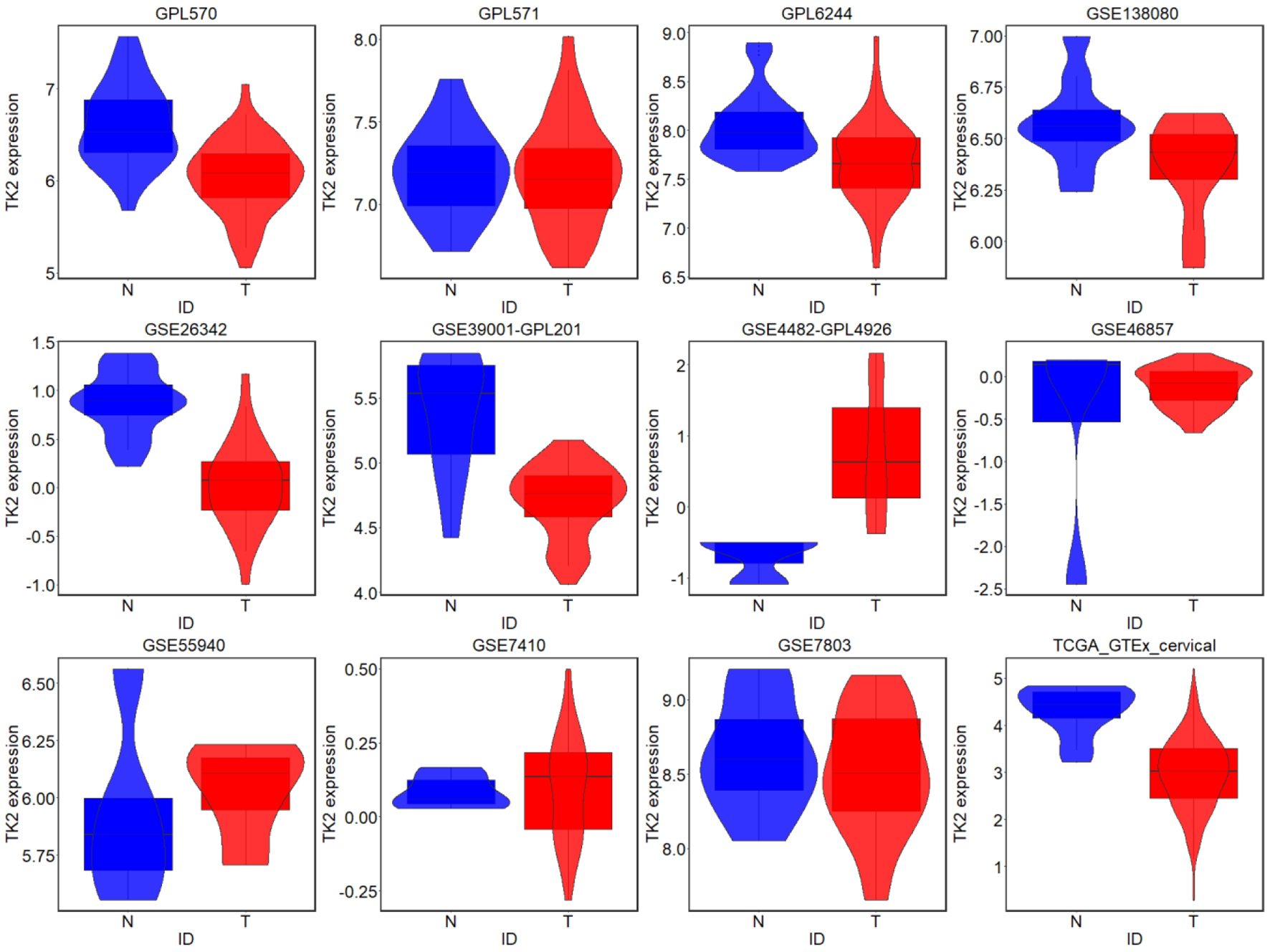
Figure 5. TK2 expression in CESC from external microarrays and RNA-seq datasets; N: non-cancer controls; T: CESC samples.
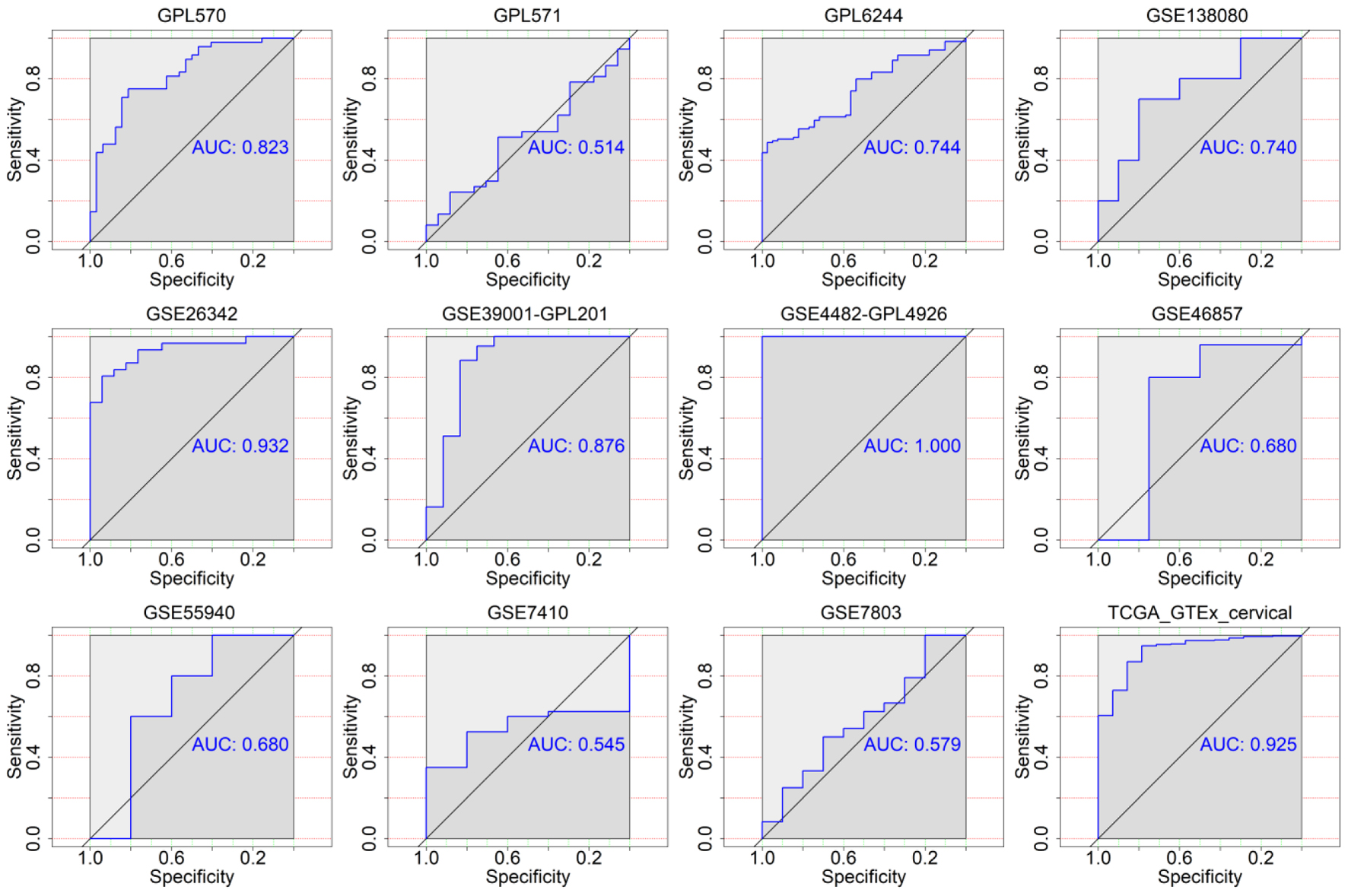
Figure 6. The discriminatory ability of TK2 expression in distinguishing CESC from non-cancer tissues in each microarray and RNA-seq dataset. AUC: area under curve.
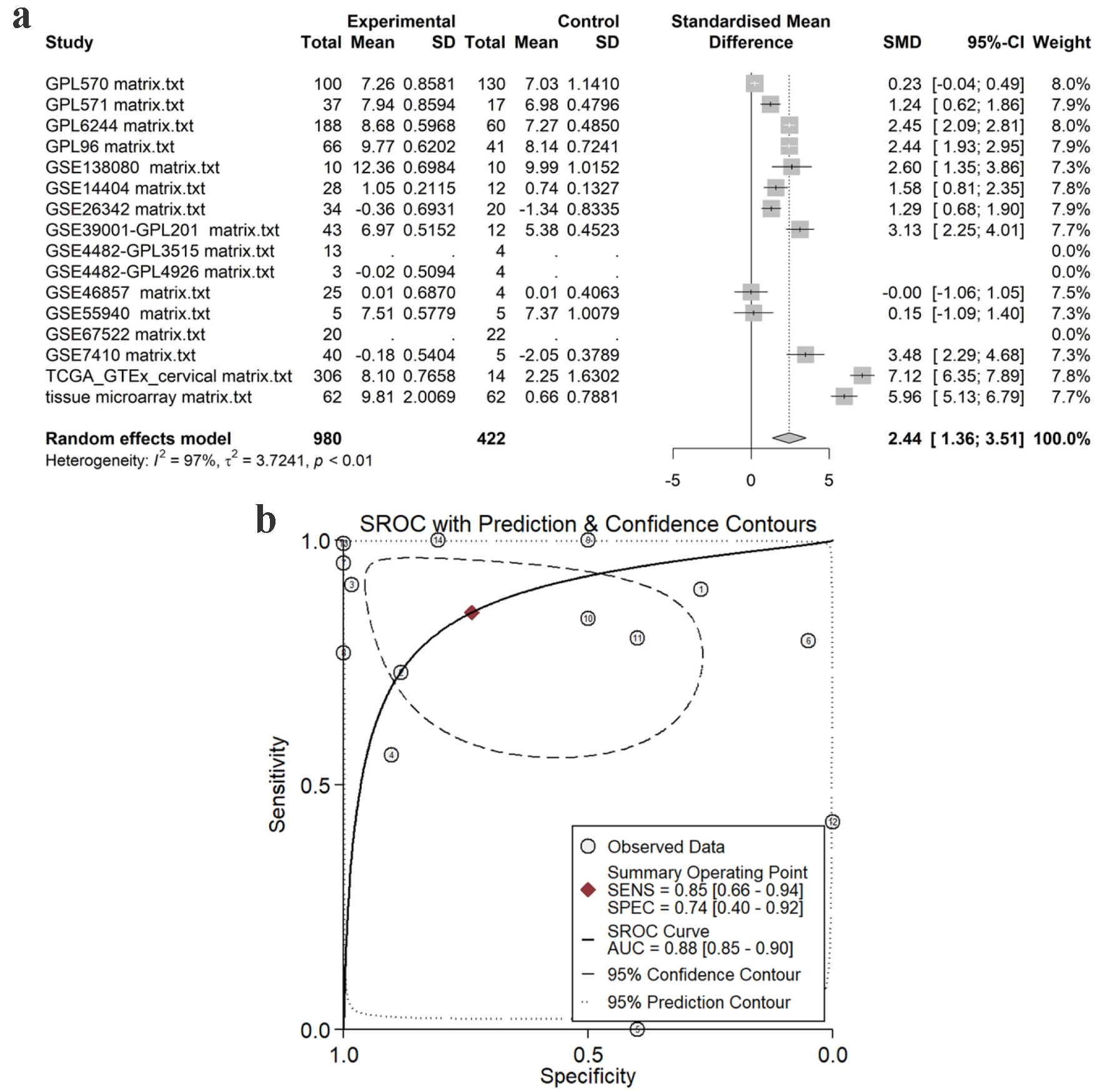
Figure 7. Pooled TK1 expression in CESC tissues. (a) SMD forest. (b) sROC curve. SMD: standardized mean difference; sROC: summarized receiver’s operating characteristics.
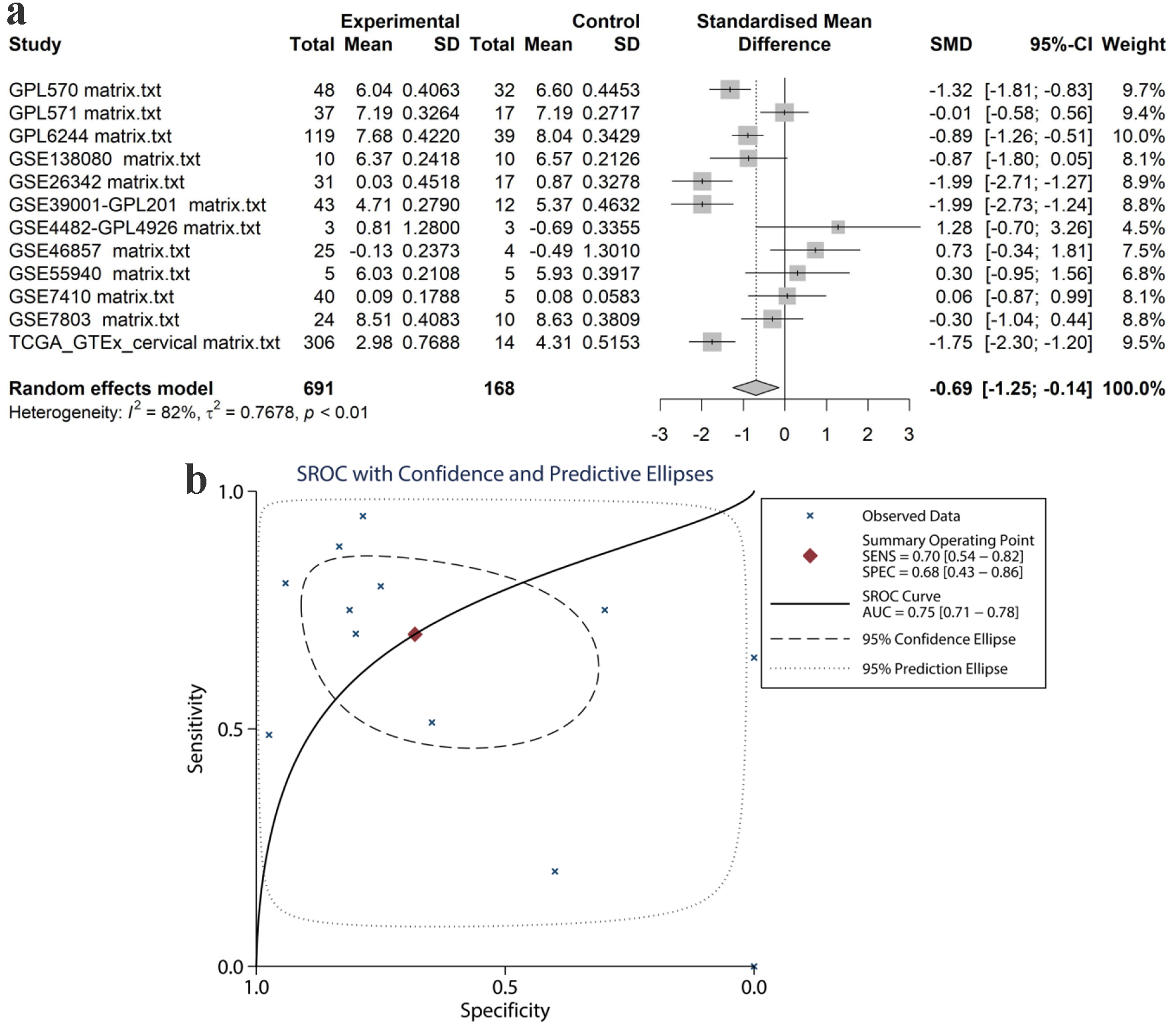
Figure 8. Pooled TK2 expression in CESC tissues. (a) SMD forest. (b) sROC curve. SMD: standardized mean difference; sROC: summarized receiver’s operating characteristics.
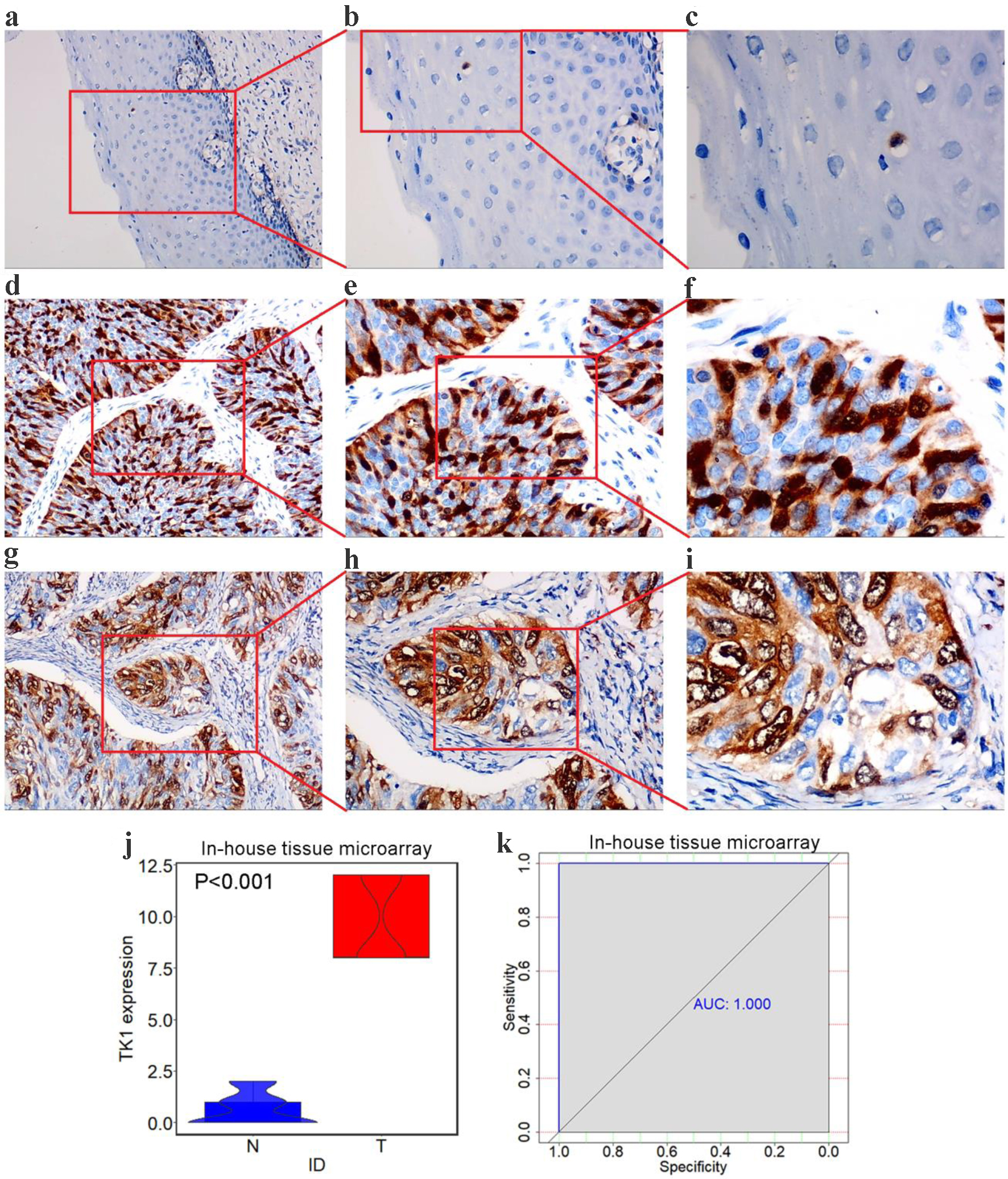
Figure 9. TK1 protein levels in CESC from tissue microarrays. (a) Negative staining of TK1 in non-cancer squamous epithelium tissues (× 100). (b) Negative staining of TK1 in non-cancer squamous epithelium tissues (× 200); (c) Negative staining of TK1 in non-cancer squamous epithelium tissues (× 400); (d, g) Strong staining of TK1 in CESC tissues (× 100); (e, h) Strong staining of TK1 in CESC tissues (× 200); (f, i) Strong staining of TK1 in CESC tissues (× 400); (j) Violin plots of TK1 expression in CESC and non-cancer controls; (k) ROC curves of the discriminating ability of TK1 overexpression. N: non-cancer samples; T: CESC samples; AUC: area under curve.
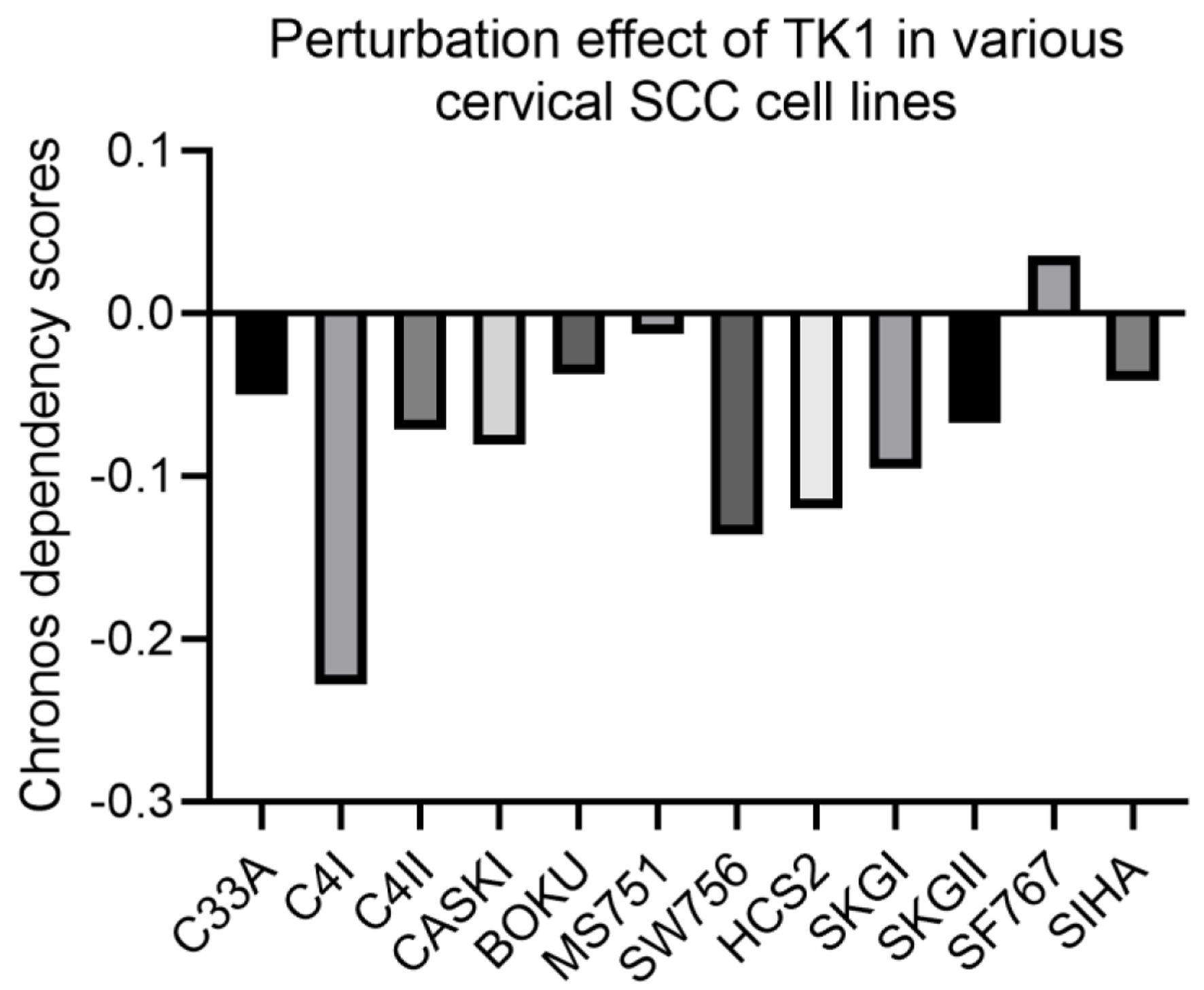
Figure 10. Perturbation effect of knocking down TK1 expression in various CESC cell lines. A lower Chronos score indicates a higher likelihood that the gene of interest is essential in a given cell line.
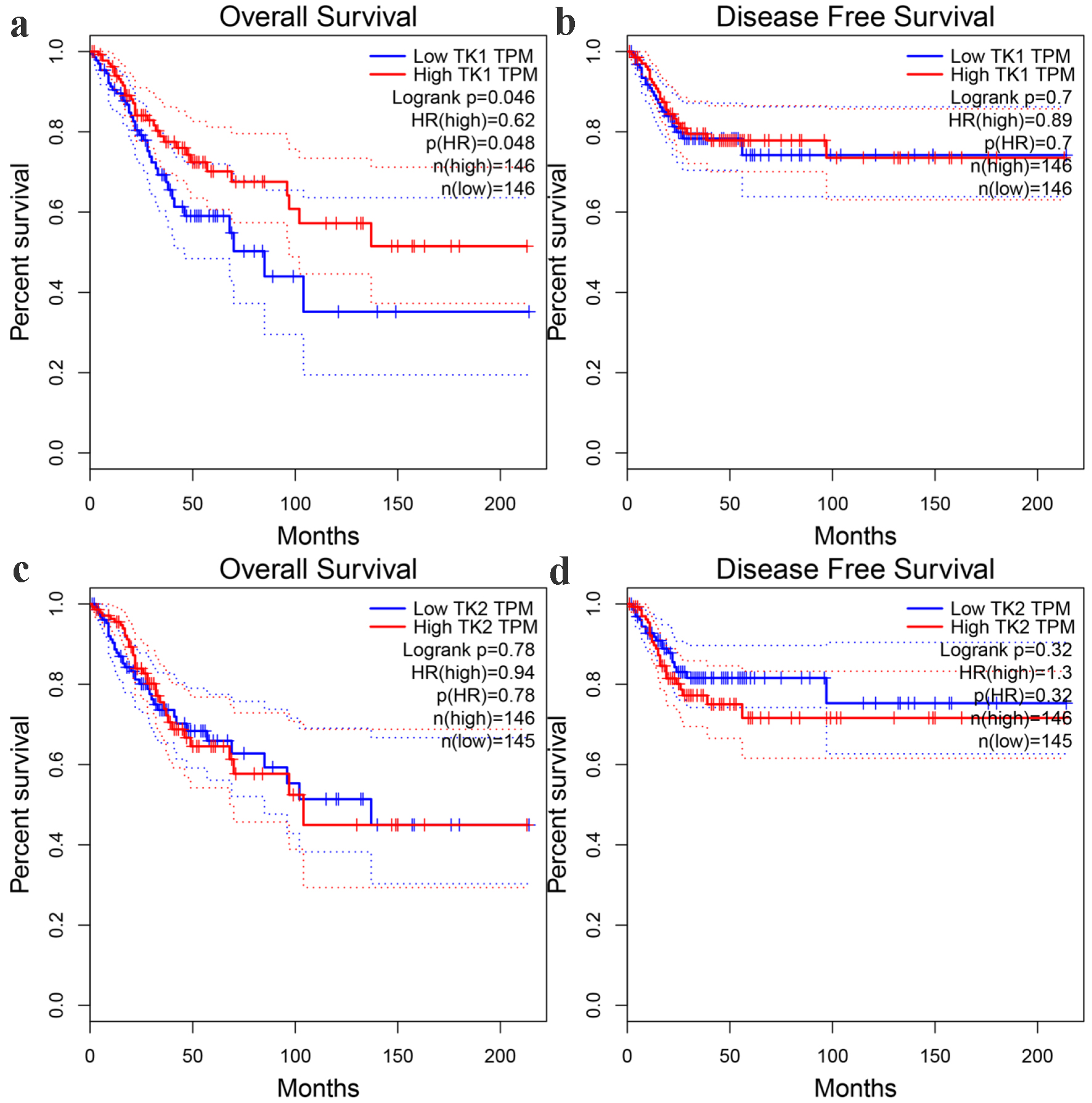
Figure 11. The prognostic significance of TK1 and TK2 expression for CESC. (a) Kaplan-Meier survival curves for overall survival of CESC patients with low or high TK1 expression. (b) Kaplan-Meier survival curves for disease-free survival of CESC patients with low or high TK1 expression. (c) Kaplan-Meier survival curves for overall survival of CESC patients with low or high TK2 expression. (d) Kaplan-Meier survival curves for disease-free survival of CESC patients with low or high TK2 expression. HR: hazard ratio.
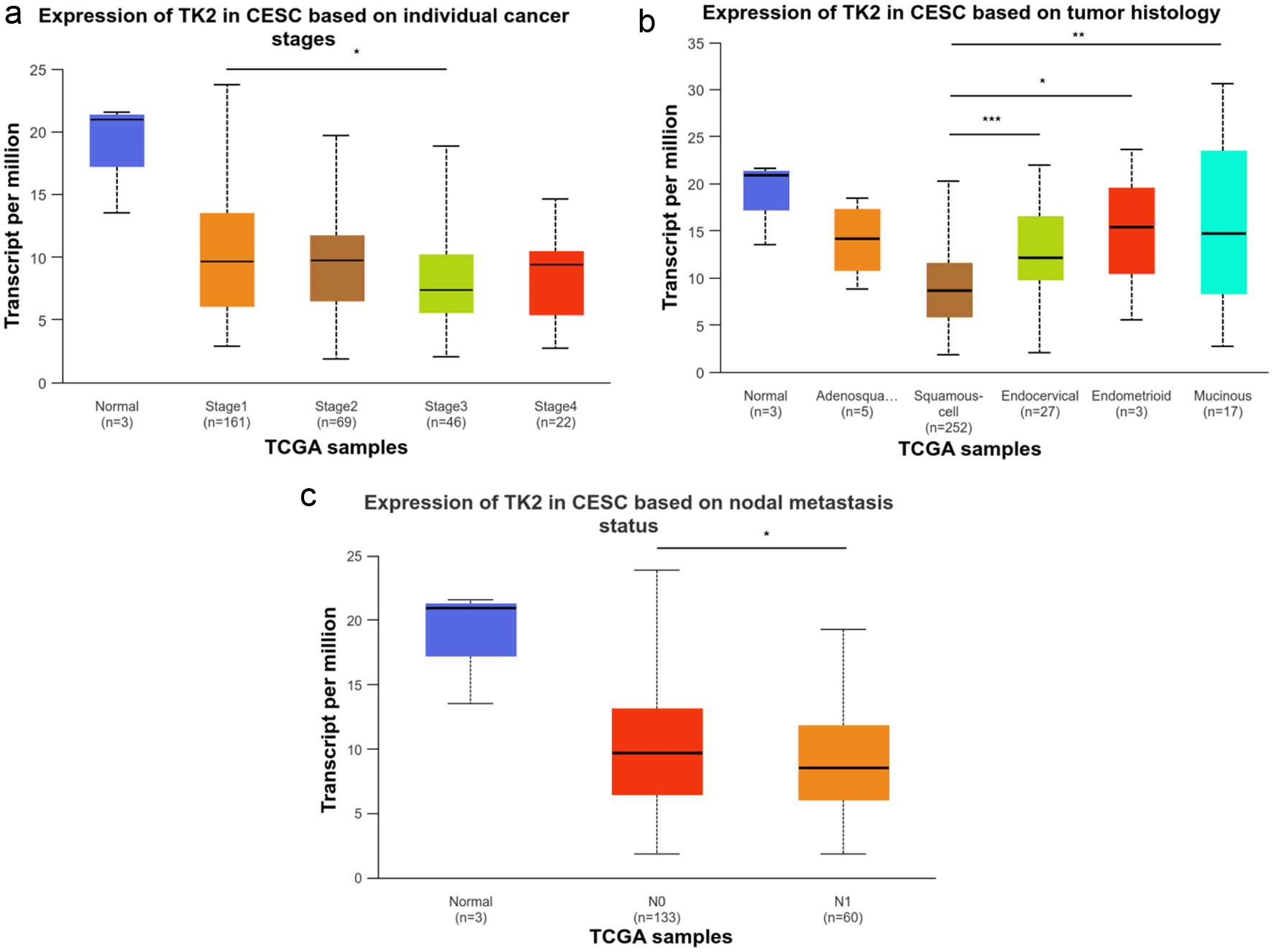
Figure 12. The relationship between TK2 expression and the clinical progression of CESC. (a) TK2 expression in CESC patients with different cancer stages. (b) TK2 expression in CESC patients with different tumor histology. (c) TK2 expression in CESC patients with different status of nodal metastasis.
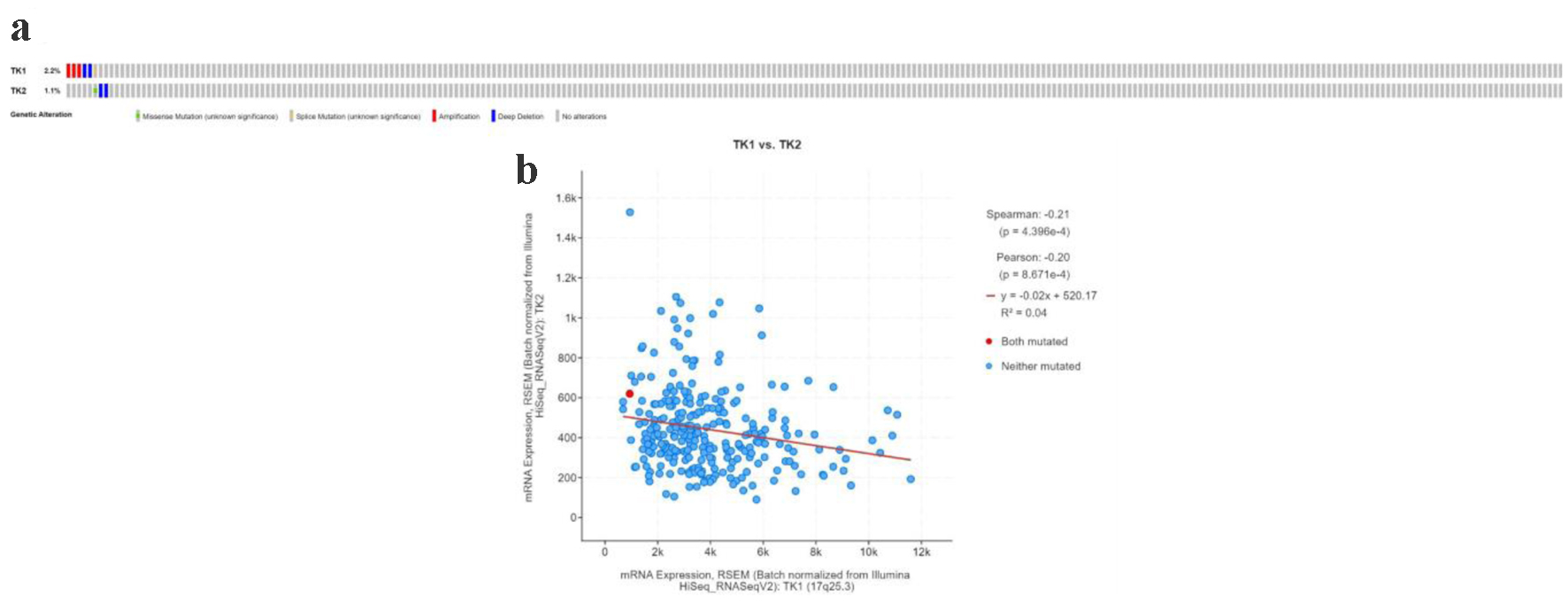
Figure 13. The TK1 and TK2 gene alterations in CESC patients. (a) The mutation type of TK1 and TK2 in CESC patients. (b) The expression of TK1 and TK2 showed a negative correlation.
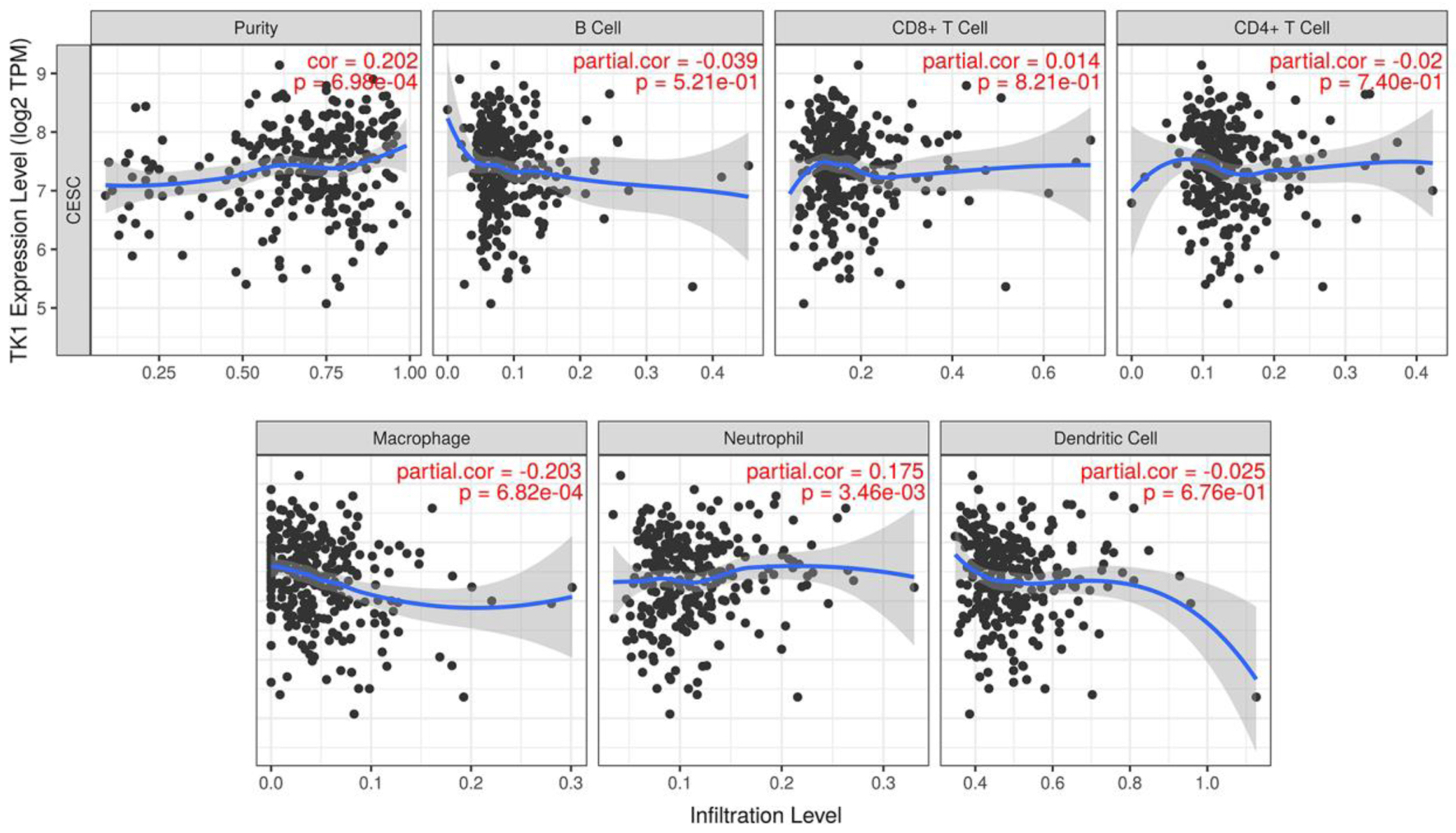
Figure 14. The correlations between TK1 expression and the infiltration level of immune cells in CESC. Scatter plot of the correlations between TK1 expression and immune infiltration. TPM: transcripts per kilobase million.
Figure 15. The correlations between TK2 expression and the infiltration level of immune cells in CESC. Scatter plot of the correlations between TK2 expression and immune infiltration. TPM: transcripts per kilobase million.
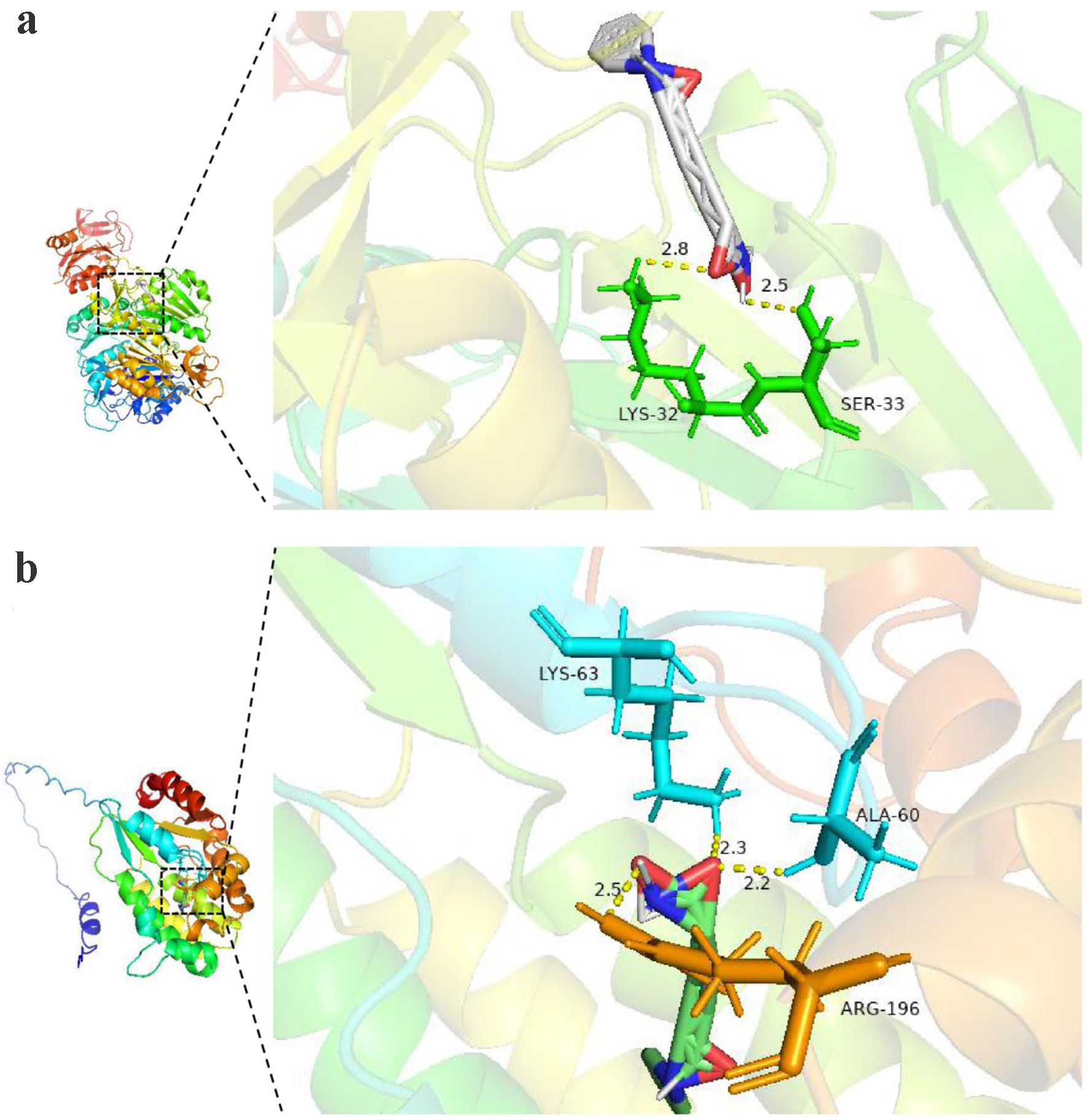
Figure 16. The molecular docking model of targeted protein and vorinostat (a: TK1 protein; b: TK2 protein).
Tables
Table 1. Basic Information From All Included RNA-seq and Microarray Datasets of Cervical Cancer
| Dataset | Platform | Country | First author | Sample type | Number of tumor samples | Number of non-cancer samples |
|---|
| GSE7803 | GPL96 | USA | Rork Kuick | Tissue | 66 | 41 |
| GSE9750 | GPL96 | USA | Murty Vundavalli | Tissue and cell lines | | |
| GSE46857 | GPL7025 | India | Rita Mulherkar | Tissue | 25 | 4 |
| GSE14404 | GPL6699 | India | Rajkumar T | Tissue | 28 | 12 |
| GSE29570 | GPL6244 | Mexico | Mariano Guardado-Estrada | Tissue | 188 | 60 |
| GSE52903 | GPL6244 | Mexico | Ingrid Medina Martinez | Tissue | | |
| GSE52904 | GPL6244 | Mexico | Ingrid Medina Martinez | Tissue | | |
| GSE89657 | GPL6244 | Mexico | Mauricio Salcedo Vargas | Tissue and cell lines | | |
| GSE39001 | GPL6244 | Mexico | Ana Maria Espinosa | Tissue | | |
| GSE27678 | GPL571 | United Kingdom | Ian Roberts | Tissue and cell lines | 37 | 17 |
| GSE63678 | GPL571 | USA | Prokopios Alexandros Polyzos | Tissue | | |
| GSE6791 | GPL570 | USA | Paul Ahlquist | Tissue | 100 | 130 |
| GSE27678 | GPL570 | United Kingdom | Ian Roberts | Tissue and cell lines | | |
| GSE63514 | GPL570 | USA | Johan den Boon | Tissue | | |
| GSE4482 | GPL4926 | India | Chandan Kumar | Tissue | 3 | 4 |
| GSE138080 | GPL4133 | Netherlands | Renske DM Steenbergen | Tissue | 10 | 10 |
| GSE4482 | GPL3515 | India | Chandan Kumar | Tissue | 13 | 4 |
| GSE39001 | GPL201 | Mexico | Ana Maria Espinosa | Tissue | 43 | 12 |
| GSE7410 | GPL1708 | Netherlands | Petra Biewenga | Tissue | 40 | 5 |
| GSE55940 | GPL16238 | China | Chen Ye | Tissue | 5 | 5 |
| GSE67522 | GPL10558 | United Kingdom | Sweta Sharma Saha | Tissue | 20 | 22 |
| GSE26342 | GPL1053/GPL1052 | USA | Natalia Shulzhenko | Tissue | 34 | 20 |
| TCGA-GTEx | - | - | - | Tissue | 306 | 14 |
Table 2. Top 10 Drugs With the Smallest Fold Changes Targeting TK1
| Drug name | P | q | Fold change | Specificity |
|---|
| Amsacrine | 1.93729 × 10-22 | 3.71892 × 10-20 | -3.14512 | 0.000296033 |
| Teniposide | 1.81285 × 10-17 | 2.69123 × 10-15 | -2.92398 | 0.000296824 |
| Tanespimycin | 2.29969 × 10-22 | 1.00411 × 10-19 | -2.8909 | 0.000137912 |
| MG-132 | 3.37601 × 10-14 | 5.36979 × 10-11 | -2.82185 | 0.000456621 |
| BRD-K68548958 | 9.27931 × 10-32 | 1.57734 × 10-29 | -2.8178 | 0.000294118 |
| SA-1919710 | 1.37796 × 10-13 | 3.83556 × 10-10 | -2.80677 | 0.001135074 |
| Torin-2 | 1.39394 × 10-17 | 6.33474 × 10-15 | -2.80657 | 0.000264831 |
| Vorinostat | 6.60819 × 10-17 | 3.75385 × 10-15 | -2.7822 | 0.000131492 |
| PP-110 | 3.48166 × 10-29 | 4.78578 × 10-27 | -2.77853 | 0.000152602 |
Table 3. Top 10 Drugs With the Smallest Fold Changes Targeting TK2
| Drug name | P | q | Fold change | Specificity |
|---|
| Trichostatin-a | 8.01858 × 10-11 | 1.16704 × 10-8 | 2.23309 | 0.000340136 |
| Trichostatin-a | 7.35718 × 10-14 | 2.08435 × 10-12 | 1.65348 | 0.000168492 |
| BRD-K82750814 | 1.72195 × 10-6 | 0.000106217 | 1.62714 | 0.000577701 |
| Trichostatin-a | 2.55848 × 10-6 | 9.05758 × 10-5 | 1.57637 | 0.000383877 |
| Trichostatin-a | 2.433 × 10-11 | 7.10065 × 10-10 | 1.47162 | 0.000194477 |
| Panobinostat | 3.75935 × 10-22 | 1.38141 × 10-20 | 1.46121 | 0.000128074 |
| Apicidin | 4.55567 × 10-19 | 1.0074 × 10-17 | 1.43291 | 0.000134174 |
| Trichostatin-a | 9.1091 × 10-11 | 1.3011 × 10-9 | 1.4319 | 0.000171028 |
| Tozasertib | 5.76929 × 10-6 | 0.000052139 | 1.41104 | 0.000120802 |
| Vorinostat | 1.72852 × 10-10 | 2.62555 × 10-9 | 1.38205 | 0.00017328 |
















