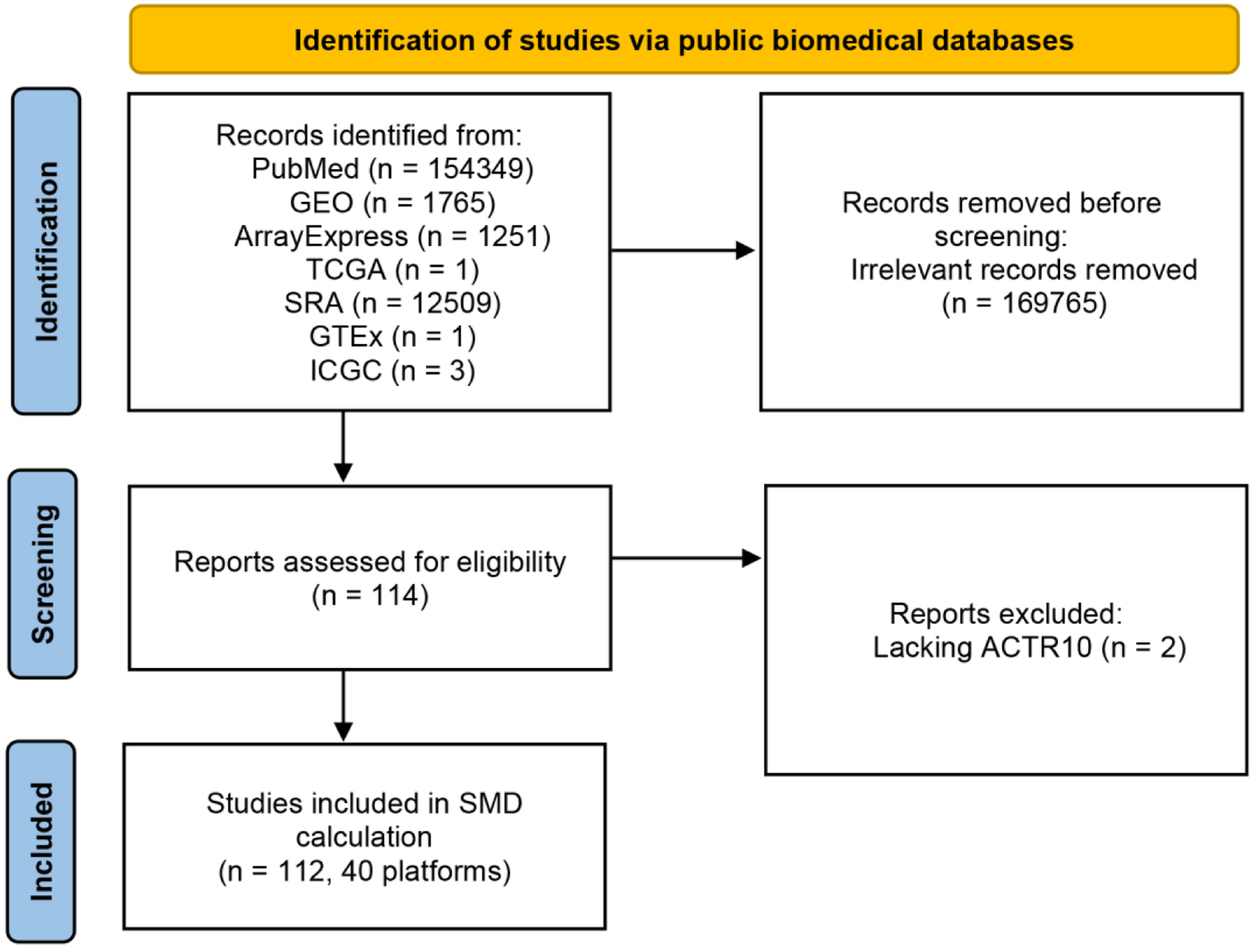
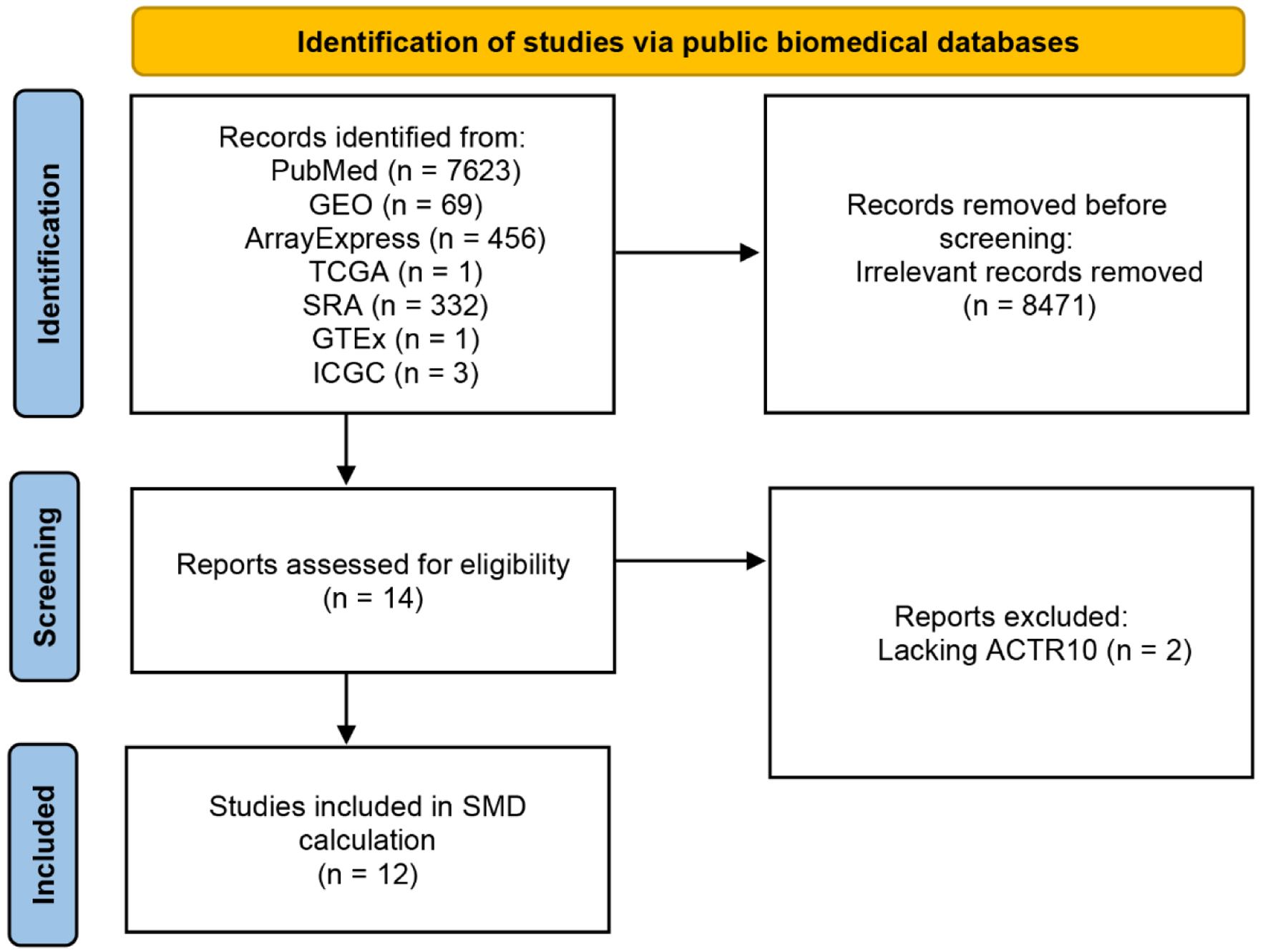
Figure 1. Flow chart of ACTR10 mRNA dataset screening. GEO: Gene Expression Omnibus; GTEx: Genotype-Tissue Expression; ICGC: International Cancer Genome Consortium; SRA: Sequence Read Archive; TCGA: The Cancer Genome Atlas.
| World Journal of Oncology, ISSN 1920-4531 print, 1920-454X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, World J Oncol and Elmer Press Inc |
| Journal website https://www.wjon.org |
Original Article
Volume 15, Number 6, December 2024, pages 882-901
ACTR10 Overexpression Facilitates the Progression and Tyrosine Kinase Inhibitor Resistance in Hepatocellular Carcinoma
Figures

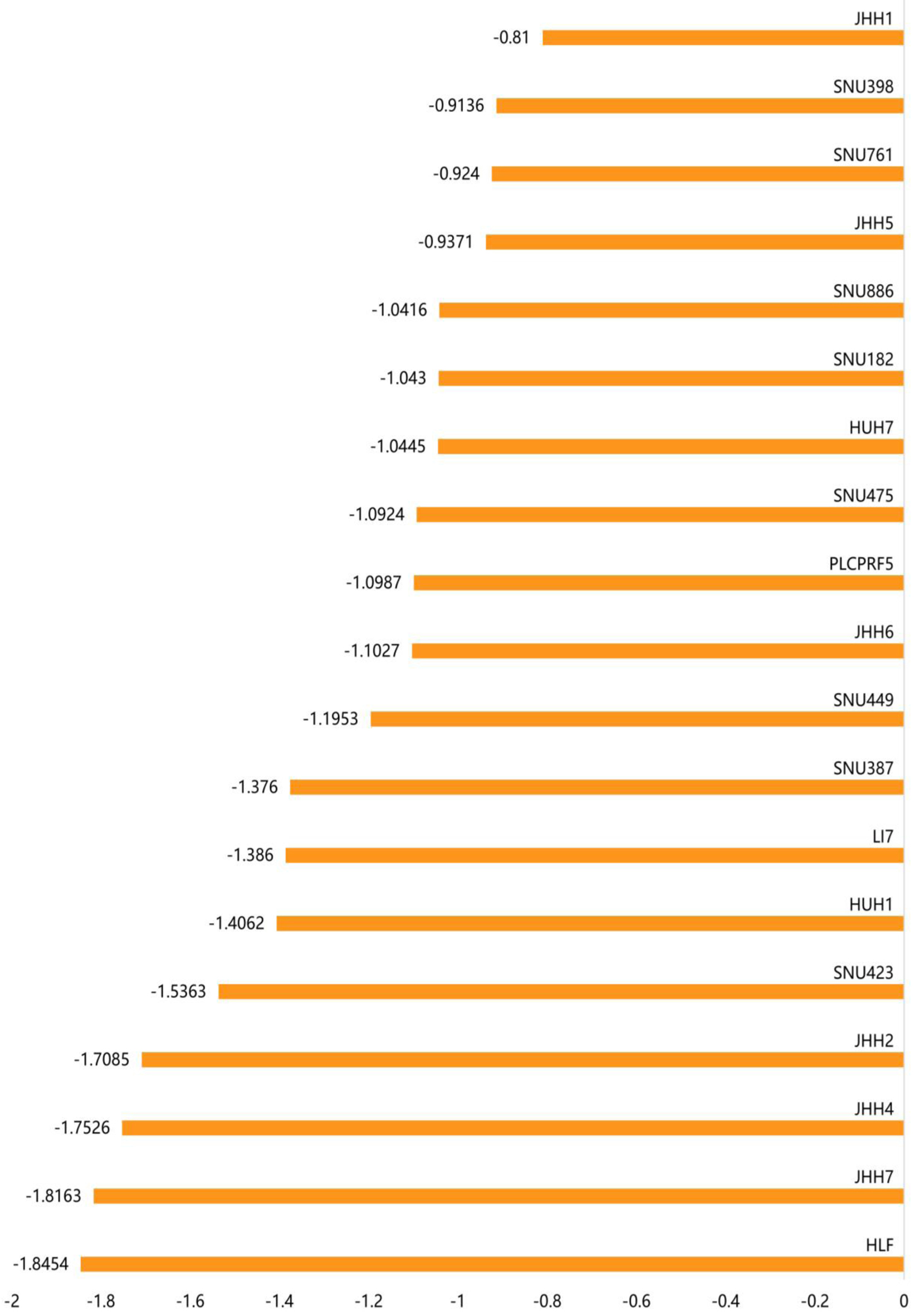
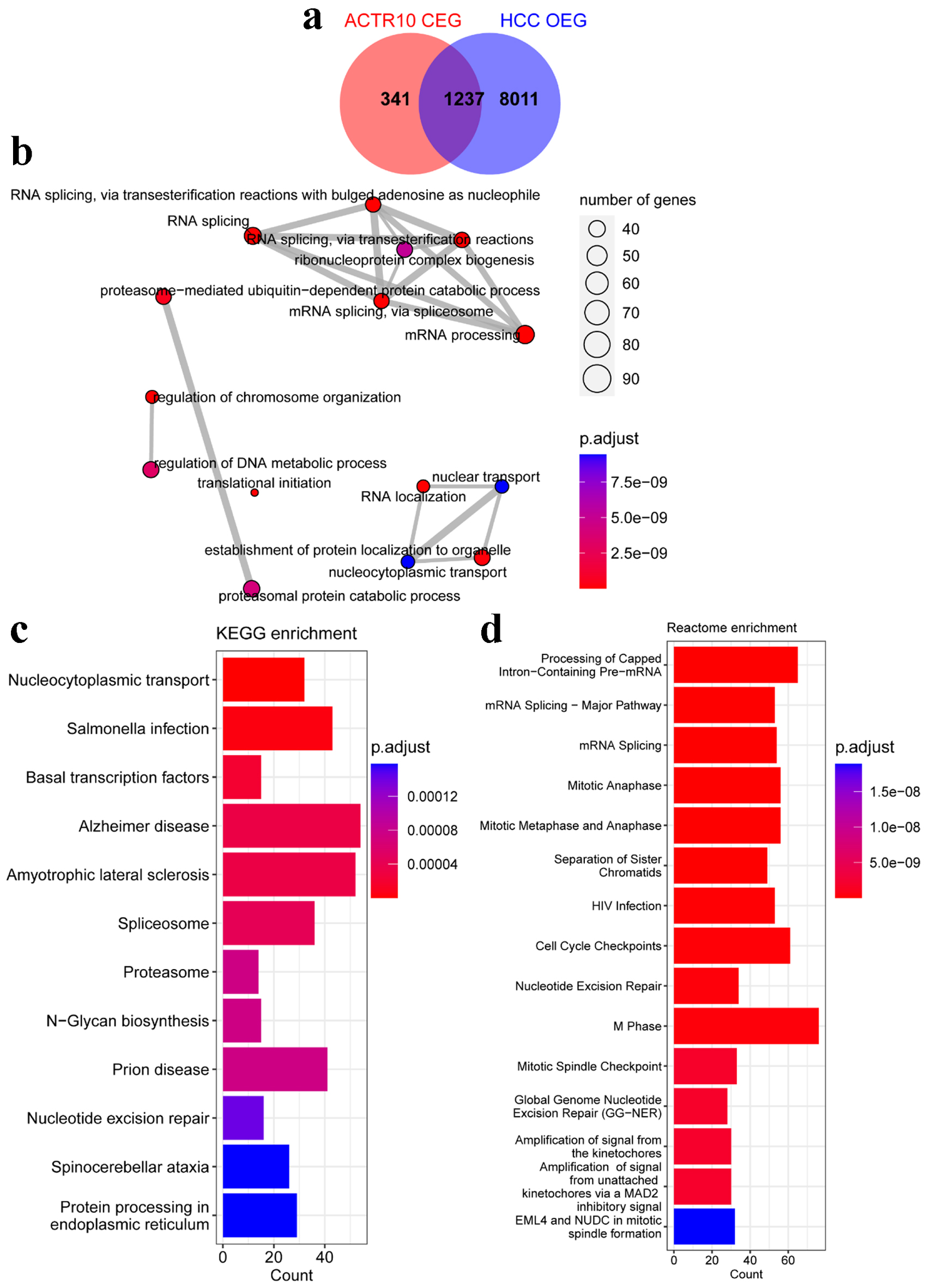
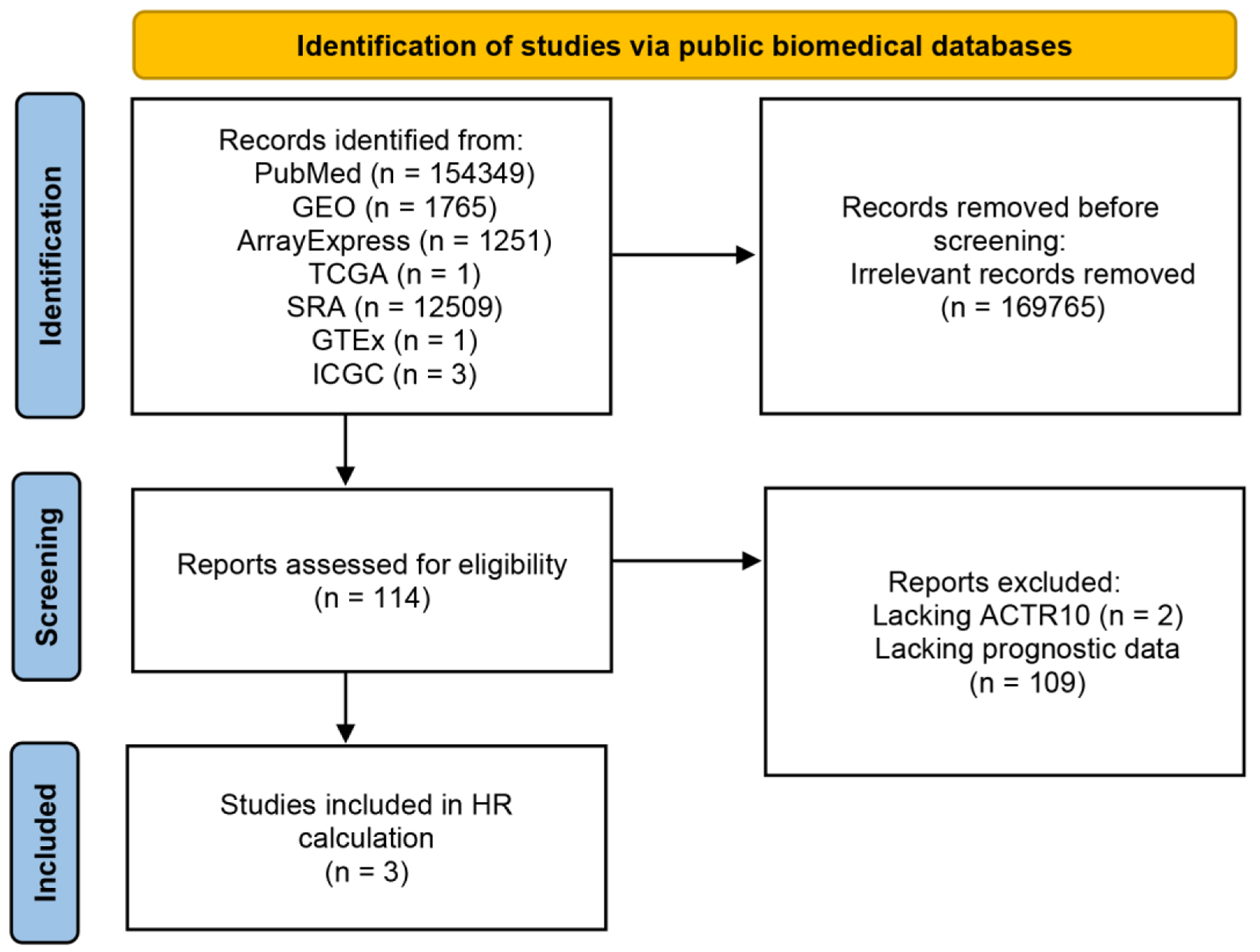
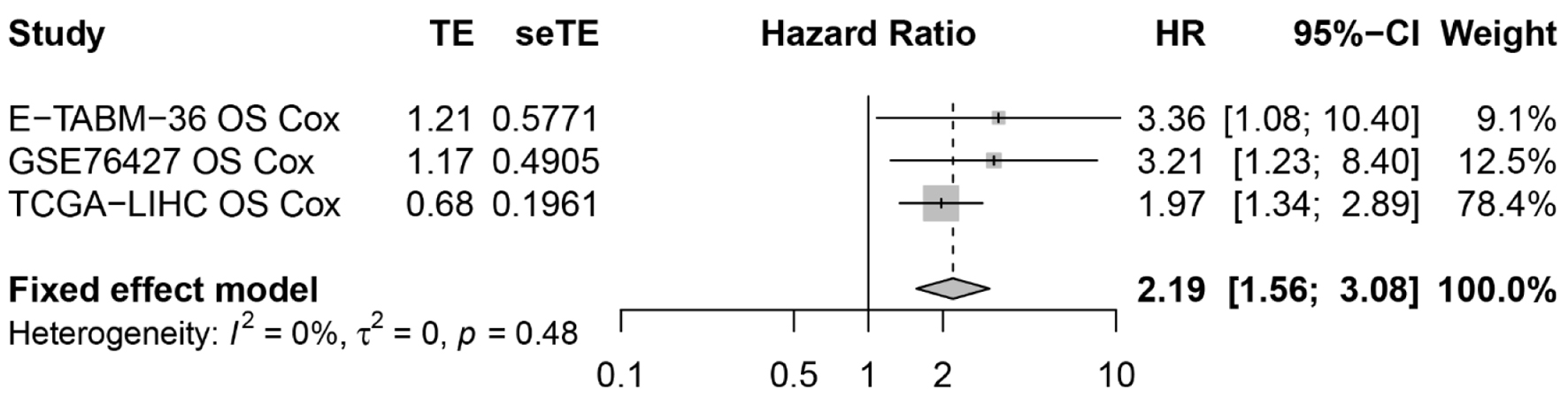
Tables
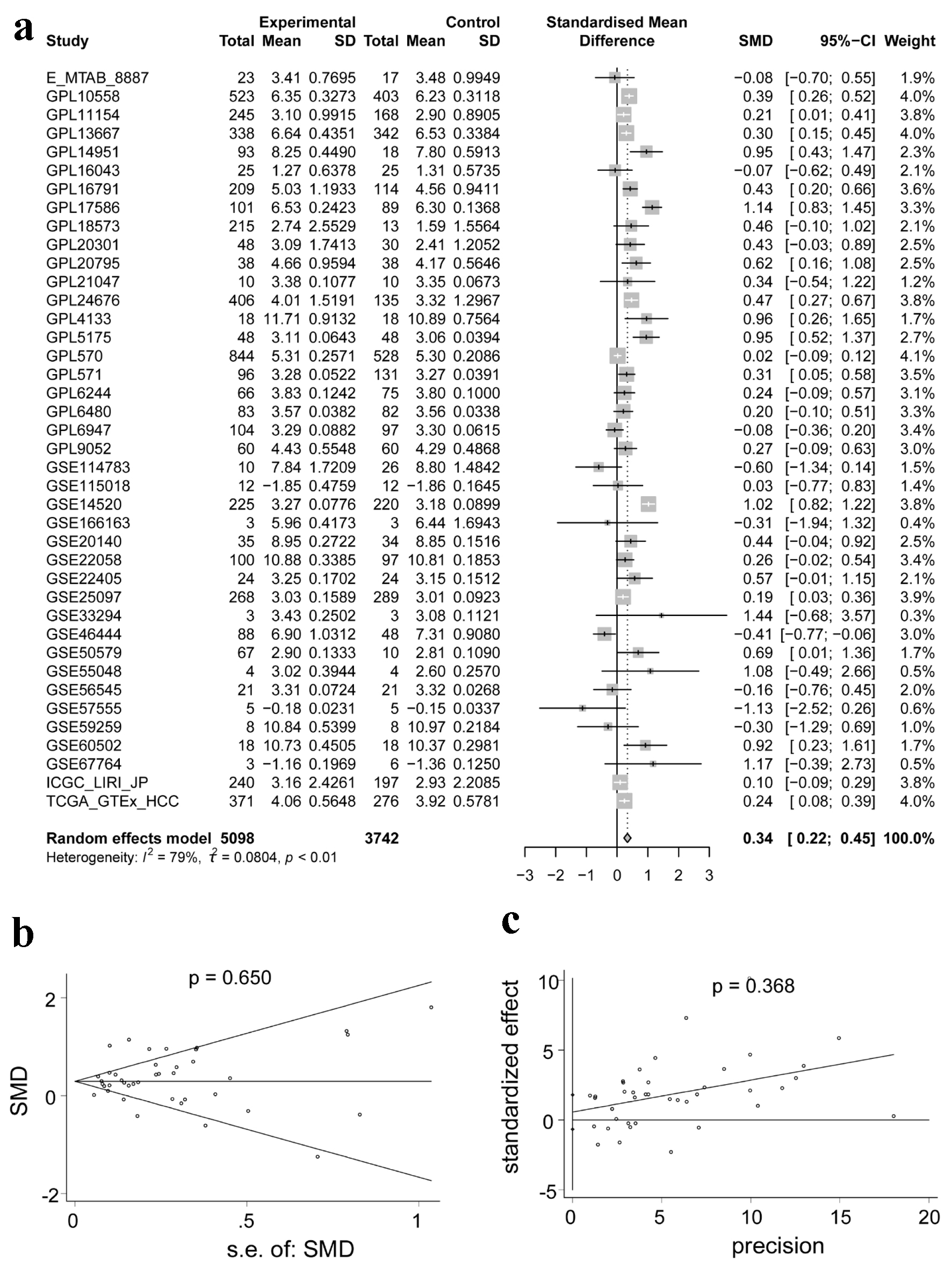
| Study | NHCC | MHCC | SDHCC | Nnon-HCC liver | Mnon-HCC liver | SDnon-HCC liver |
|---|---|---|---|---|---|---|
| NHCC: the number of HCC samples; MHCC: the mean expression level of ACTR10 in HCC samples; SDHCC: the standard deviation of ACTR10 expression in HCC samples; Nnon-HCC liver: the number of non-HCC liver samples; Mnon-HCC liver: the mean expression level of ACTR10 in non-HCC liver samples; SDnon-HCC liver: the standard deviation of ACTR10 expression in non-HCC liver samples; HCC: hepatocellular carcinoma. | ||||||
| E_MTAB_8887 | 23 | 3.41 | 0.77 | 17 | 3.48 | 0.99 |
| GPL10558 | 523 | 6.35 | 0.33 | 403 | 6.23 | 0.31 |
| GPL11154 | 245 | 3.10 | 0.99 | 168 | 2.90 | 0.89 |
| GPL13667 | 338 | 6.64 | 0.44 | 342 | 6.53 | 0.34 |
| GPL14951 | 93 | 8.25 | 0.45 | 18 | 7.80 | 0.59 |
| GPL16043 | 25 | 1.27 | 0.64 | 25 | 1.31 | 0.57 |
| GPL16791 | 209 | 5.03 | 1.19 | 114 | 4.56 | 0.94 |
| GPL17586 | 101 | 6.53 | 0.24 | 89 | 6.30 | 0.14 |
| GPL18573 | 215 | 2.74 | 2.55 | 13 | 1.59 | 1.56 |
| GPL20301 | 48 | 3.09 | 1.74 | 30 | 2.41 | 1.21 |
| GPL20795 | 38 | 4.66 | 0.96 | 38 | 4.17 | 0.56 |
| GPL21047 | 10 | 3.38 | 0.11 | 10 | 3.35 | 0.07 |
| GPL24676 | 406 | 4.01 | 1.52 | 135 | 3.32 | 1.30 |
| GPL4133 | 18 | 11.71 | 0.91 | 18 | 10.89 | 0.76 |
| GPL5175 | 48 | 3.11 | 0.06 | 48 | 3.06 | 0.04 |
| GPL570 | 844 | 5.31 | 0.26 | 528 | 5.30 | 0.21 |
| GPL571 | 96 | 3.28 | 0.05 | 131 | 3.27 | 0.04 |
| GPL6244 | 66 | 3.83 | 0.12 | 75 | 3.80 | 0.10 |
| GPL6480 | 83 | 3.57 | 0.04 | 82 | 3.56 | 0.03 |
| GPL6947 | 104 | 3.29 | 0.09 | 97 | 3.30 | 0.06 |
| GPL9052 | 60 | 4.43 | 0.55 | 60 | 4.29 | 0.49 |
| GSE114783 | 10 | 7.84 | 1.72 | 26 | 8.80 | 1.48 |
| GSE115018 | 12 | -1.85 | 0.48 | 12 | -1.86 | 0.16 |
| GSE14520 | 225 | 3.27 | 0.08 | 220 | 3.18 | 0.09 |
| GSE166163 | 3 | 5.96 | 0.42 | 3 | 6.44 | 1.69 |
| GSE20140 | 35 | 8.95 | 0.27 | 34 | 8.85 | 0.15 |
| GSE22058 | 100 | 10.88 | 0.34 | 97 | 10.81 | 0.19 |
| GSE22405 | 24 | 3.25 | 0.17 | 24 | 3.15 | 0.15 |
| GSE25097 | 268 | 3.03 | 0.16 | 289 | 3.01 | 0.09 |
| GSE33294 | 3 | 3.43 | 0.25 | 3 | 3.08 | 0.11 |
| GSE46444 | 88 | 6.90 | 1.03 | 48 | 7.31 | 0.91 |
| GSE50579 | 67 | 2.90 | 0.13 | 10 | 2.81 | 0.11 |
| GSE55048 | 4 | 3.02 | 0.39 | 4 | 2.60 | 0.26 |
| GSE56545 | 21 | 3.31 | 0.07 | 21 | 3.32 | 0.03 |
| GSE57555 | 5 | -0.18 | 0.02 | 5 | -0.15 | 0.03 |
| GSE59259 | 8 | 10.84 | 0.54 | 8 | 10.97 | 0.22 |
| GSE60502 | 18 | 10.73 | 0.45 | 18 | 10.37 | 0.30 |
| GSE67764 | 3 | -1.16 | 0.20 | 6 | -1.36 | 0.12 |
| ICGC_LIRI_JP | 240 | 3.16 | 2.43 | 197 | 2.93 | 2.21 |
| TCGA_GTEx_HCC | 371 | 4.06 | 0.56 | 276 | 3.92 | 0.58 |
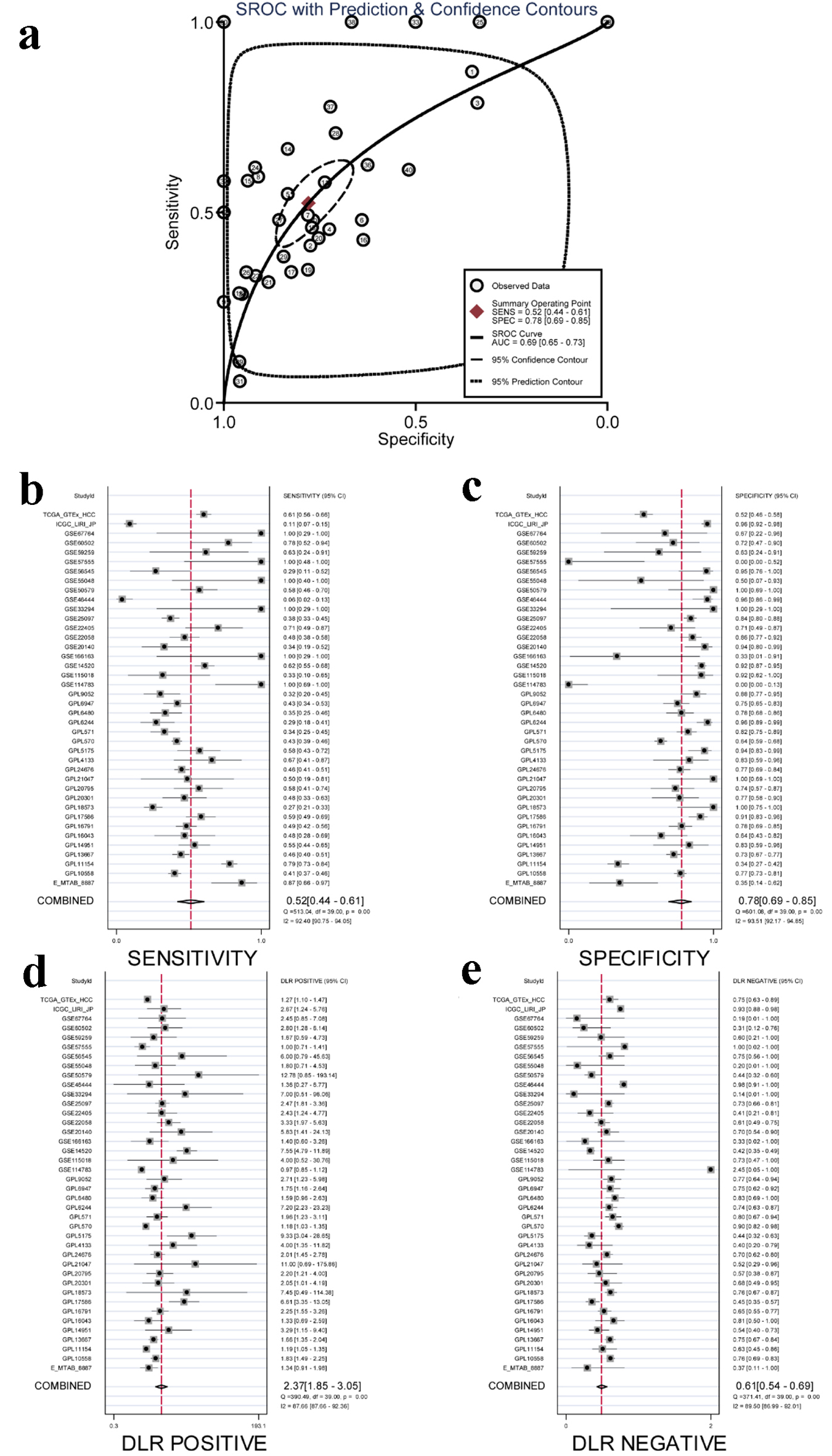
| Study | HCC | Platform |
|---|---|---|
| HCC: hepatocellular carcinoma. | ||
| E-TABM-36 | 41 | Affymetrix HG-U133A GeneChips arrays |
| GSE76427 | 90 | GPL10558 |
| TCGA-LIHC | 321 | RNA-seq |
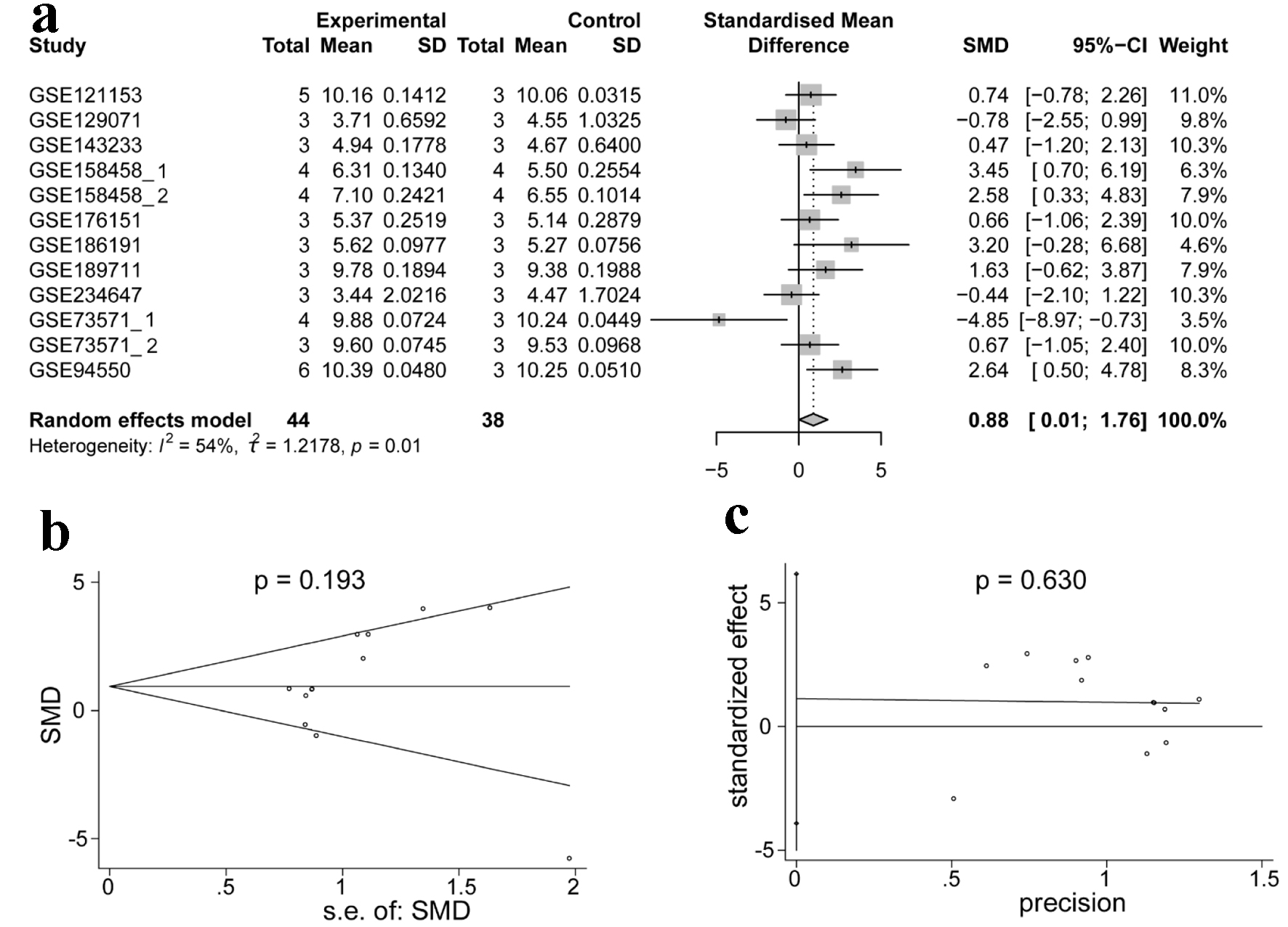
| Study | NTKI-S | MTKI-S | SDTKI-S | NTKI-R | MTKI-R | SDTKI-R |
|---|---|---|---|---|---|---|
| TKI: tyrosine kinase inhibitor; TKI-S: TKI-sensitive; TKI-R: TKI-resistant. | ||||||
| GSE121153 | 5 | 10.16 | 0.14 | 3 | 10.06 | 0.03 |
| GSE129071 | 3 | 3.71 | 0.66 | 3 | 4.55 | 1.03 |
| GSE143233 | 3 | 4.94 | 0.18 | 3 | 4.67 | 0.64 |
| GSE158458_1 | 4 | 6.31 | 0.13 | 4 | 5.50 | 0.26 |
| GSE158458_2 | 4 | 7.10 | 0.24 | 4 | 6.55 | 0.10 |
| GSE176151 | 3 | 5.37 | 0.25 | 3 | 5.14 | 0.29 |
| GSE186191 | 3 | 5.62 | 0.10 | 3 | 5.27 | 0.08 |
| GSE189711 | 3 | 9.78 | 0.19 | 3 | 9.38 | 0.20 |
| GSE234647 | 3 | 3.44 | 2.02 | 3 | 4.47 | 1.70 |
| GSE73571_1 | 4 | 9.88 | 0.07 | 3 | 10.24 | 0.04 |
| GSE73571_2 | 3 | 9.60 | 0.07 | 3 | 9.53 | 0.10 |
| GSE94550 | 6 | 10.39 | 0.05 | 3 | 10.25 | 0.05 |
| Cell line | Time | Dose | P-value | q-value | logFoldChange | Specificity | |
|---|---|---|---|---|---|---|---|
| DGB: Drug Gene Budger. | |||||||
| Trichostatin A | MCF7 | 6 h | 0.1 µM | 0 | 0 | -1.35095 | 0.000249563 |
| Trichostatin A | HL60 | 6 h | 0.1 µM | 5.33468 × 10-38 | 5.22588×10-36 | -1.07017 | 0.000342466 |
| Trichostatin A | HL60 | 6 h | 1.0 µM | 1.35366 × 10-13 | 1.00243×10-11 | -1.74451 | 0.000390778 |